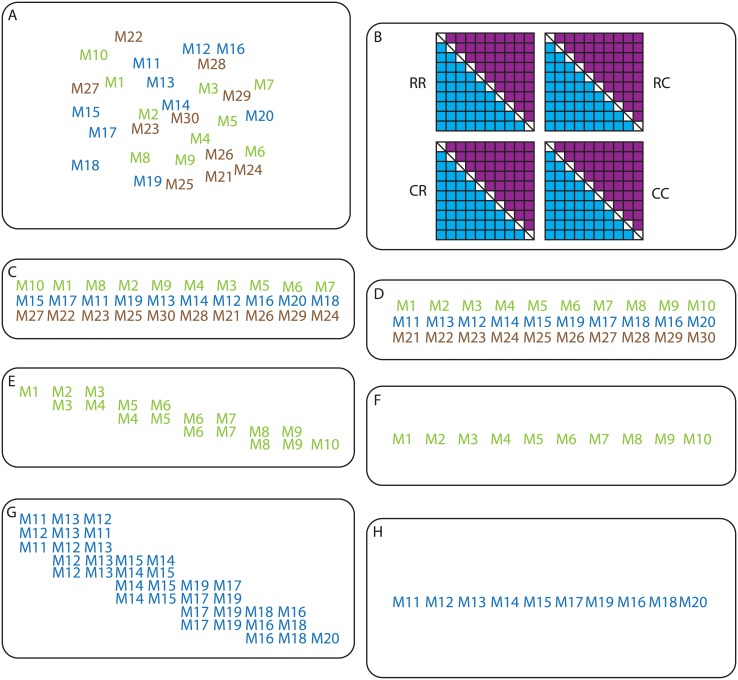
Fig 1. An overview of the BatchMap pipeline.
A) Input data file is read into BatchMap using the function read.outcross2(). B) A two-point table of recombination fractions is created and likelihoods calculated per phase using the function rf.2pts(). C) Markers are assigned to linkage groups (LGs) using the function group(), sequence objects created using make.seq() and LGs are split into parental maps using marker segregation type. D) Marker order is determined using RECORD, with the number of iterations and threads set using the function record.parallel(). E-F) The genetic map is calculated in centiMorgans by first using the function pick.batch.sizes() to split the LG into even groups and then using function map.overlapping.batches() with the predefined batch size. All pairwise marker phases are set within each batch and overlapping markers are fixed in phase from the previous batch. When all marker pairs are fixed in phase the original OneMap function map() is called to calculate the distance between markers in cM using either the Kosambi or Haldane mapping function. G-H) An optional ripple function, ripple.ord(), can be set in map.overlapping.batches() to consider all single pairwise swaps between markers in a sliding window of predefined size along the LG. When the best order (based on likelihoods) within each window is set, map calculation will be carried out as in E-F).