Abstract
The single-stranded DNA (ssDNA) cytidine deaminase APOBEC3F (A3F) deaminates cytosine (C) to uracil (U) and is a known restriction factor of HIV-1. Its C-terminal catalytic domain (CD2) alone is capable of binding single-stranded nucleic acids and is important for deamination. However, little is known about how the CD2 interacts with single-stranded DNA (ssDNA). Here we report a crystal structure of A3F-CD2 in complex with a 10 nucleotide (nt) ssDNA composed of poly-thymine, which reveals a novel positively-charged nucleic acid binding site distal to the active center that plays a key role in substrate DNA binding and catalytic activity. Lysine and tyrosine residues within this binding site interact with the ssDNA, and mutating these residues dramatically impairs both ssDNA binding and catalytic activity. This binding site is not conserved in APOBEC3G (A3G), which may explain differences in ssDNA binding characteristics between A3F-CD2 and A3G-CD2. In addition, we observed an alternative Zn-coordination conformation around the active center. These findings reveal the structural relationships between nucleic acid interactions and catalytic activity of A3F.
Graphical abstract
Introduction
The members of the APOBEC family of cytidine deaminases share a conserved zinc-coordinating cytidine deaminase domain (CD) and generally possess the ability to catalyze cytosine into uracil on ssDNA or RNA. In humans, there are eleven related members that fall within the APOBEC family: APOBEC-1, APOBEC-2, seven APOBEC-3s (A3A through A3H), APOBEC-4, and Activation-Induced Deaminase (AID). These are involved in diverse biological processes including lipid metabolism, antibody generation, virus/retroelement restriction, and cancer genome hypermutation (reviewed and references therein(1–4)). The A3 subfamily is particularly well known for being a key part of the innate immune response against viral infection and intrinsic retroelements (reviewed and references therein(5–8)). The double-domain A3F and A3G are among the A3 proteins that have been characterized as potent antiviral factors and can restrict HIV-1 replication in the absence of HIV-1 Vif (9–13). A3F and A3G can be incorporated into HIV virions, inhibiting HIV-1 replication by blocking reverse transcription, introducing dC-to-dU hypermutations in viral cDNA (or G-to-A hypermutations in viral DNA), and inhibiting proviral DNA formation(9,10,13–21). However, the HIV viral protein Vif suppresses A3F, A3G and some other APOBEC proteins by mediating their degradation through the Cul5-E3 ligase pathway(22–29).
A3F and A3G both contain two CD domains. Although the C-terminal CD2 of both A3F and A3G are responsible for catalysis, A3F-CD2 and A3G-CD2 display distinctive binding for substrate ssDNA(30–32). The catalytic activity of A3G-CD2 alone is about three orders of magnitude weaker than full-length A3G, and the ssDNA binding of A3G-CD2 is barely detectable by a gel-shift assay(30,32,33). In contrast to A3G, the activity of A3F-CD2 alone is only about 25 times weaker than full-length A3F, the ssDNA binding of A3F-CD2 alone is readily detectable by a gel-shift assay, and the presence of its CD1 domain enhances the strength of ssDNA binding as well as deamination activity by about 10 fold(31). Also, the A3F-CD2 residues on a loop close to the active center (loop 7) are shown to be involved in both ssDNA binding and catalytic activity in A3F-CD2(31). However, it is not clear how loop 7 affects the ssDNA binding, or how other structural elements outside of loop 7 away from the active center are involved in ssDNA binding.
Previous research has suggested that the CD1 domains of A3F and A3G are primarily involved in viral incorporation and nucleic acid binding, while the CD2 domain largely determines the catalytic activity and substrate specificity(31,33–43). However, it has been shown that the CD2 domains of A3F and A3G are also involved in nucleic acid binding and viral incorporation(33,37,44,45), and so an exact picture of how these functions are regulated and occur has yet to be fully explained. It may be possible that different functions may incorporate binding at multiple sites on different APOBEC proteins. For example, a recent study of mass spectrometry of A3G peptides cross-linked to bound nucleic acids revealed three separate regions on A3G involved with DNA interactions(46), each of which may independently or cooperatively bind ssDNA or RNA. Despite the publication of several apo-structures of APOBEC proteins(30,32,47–59), the only reported co-crystal structures of nucleic acids bound to members of the APOBEC family include a structure revealing ssDNA bound at the inactive Zn-center of a primate A3G-CD1(32), and structures showing ssDNA bound at the active Zn-centers of A3A and a chimeric A3B-CD2(60,61), all of which present only 3–6 nt bound to the Zn-center despite oligomers with lengths of 7–15 nt being used during crystallization. Because longer oligomeric ssDNA with lengths greater than six nucleotides generally show tighter binding and higher deamination activity(18,50,62), it is likely that substrate ssDNA must also bind to APOBEC proteins beyond the Zn-center for efficient deamination, and possibly for other biological processes. The molecular details of how nucleic acids bind to the areas outside the Zn-center is unknown for any APOBEC protein so far, including the CD2 of A3F.
Here we report a crystal structure of a wild-type CD2 of A3F in complex with a 10 nt poly-dT ssDNA that reveals an unexpected ssDNA-binding surface of A3F-CD2 that is distal to the zinc active center. Multiple tyrosine and lysine residues within the interface form hydrophobic and electrostatic interactions with the ssDNA respectively. Subsequent mutagenesis studies demonstrated that these residues on the DNA-binding surface play a role not only in ssDNA binding, but also in RNA binding and in catalytic activity. Most of these residues interacting with the ssDNA on this positively charged surface are specific to A3F-CD2 and not conserved in A3G-CD2, suggesting this ssDNA interface is a unique property of A3F-CD2. Based on this structure, a model of how A3F-CD2 interacts with its ssDNA substrate has been proposed.
Results
Crystal structure reveals a positively-charged patch of A3F-CD2 binds to a poly-dT ssDNA
In order to get the complex structure of deaminase active A3F-CD2 domain with ssDNA, we expressed and purified wild-type (WT) hA3F-CD2 (residues 190–373) that was cleaved from a GST-fusion construct. We obtained co-crystals of the WT A3F-CD2 in complex with a 10 nt poly-dT ssDNA, and determined the protein-ssDNA complex structure at a resolution of 3.7 Å. In one asymmetric unit (ASU), there are eight A3F-CD2 molecules and two well-ordered 10 nt ssDNA molecules, with each ssDNA interacting with four A3F-CD2 subunits (Table 1, Figure 1A). Each A3F-CD2 molecule contains a highly positively charged surface area (Figure 1B), composed of five lysine residues (residues 334, 337, 352, 355, and 358). This positively charged surface interacts with the negatively charged ssDNA phosphate backbone via electrostatic interactions mostly through three lysine residues K352 (K1), K355 (K2), and K358 (K3) (Figure 1C, 1D). However, two tyrosine residues located within this positively-charged surface also interact with ssDNA. Specifically, Y333 (Y1) of all four A3F-CD2 subunits form hydrophobic pi stacking interactions with the thymine bases of T1, T3, T7, T9 respectively (Figure 1C, 1D). The respective Y359 (Y2) residues on two of the A3F-CD2 subunits (chains A and D) stack with the bases of T2 and T8, whereas the other two Y359 residues (chains B and C) may interact with T4 and T6 only (Figure 1C, 1D). Therefore, the poly-dT ssDNA interacts with A3F-CD2 through a combination of electrostatic interactions with its negatively charged backbone and hydrophobic interactions with its non-paired bases, which are consistent with interactions expected for non-sequence specific binding of single stranded nucleic acids.
Table 1.
Crystallographic data collection and refinement statistics
| A3F-CD2 + 10nt ssDNA | |
|---|---|
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 68.194 108.489 108.549 |
| α, β, γ (Å) | 79.602 71.798 71.750 |
| Resolution (Å) | 50–3.70 (3.83–3.70)* |
| Rsym or Rmerge | 22.7 (94.5) |
| I/σI | 4.7 (1.1) |
| Completeness (%) | 96.2 (85.7) |
| Redundancy | 2.8 (2.4) |
| Molecules per ASU | 10 |
| Refinement | |
| Resolution (Å) | 50–3.70 |
| No. reflections | 28753 |
| Rwork/Rfree | 24.98/26.66 |
| No. atoms | 12926 |
| Protein | 12520 |
| Ligand/ion | 406 |
| B-factors | 107.9 |
| Protein | 107.6 |
| Ligand/ion | 121.9 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.1 |
| Ramachandran plot | |
| Favored | 81.2% |
| Allowed | 16.1% |
| Outlier | 2.7% |
Structure was determined from a single crystal.
Highest-resolution shell is shown in parentheses.
Figure 1.
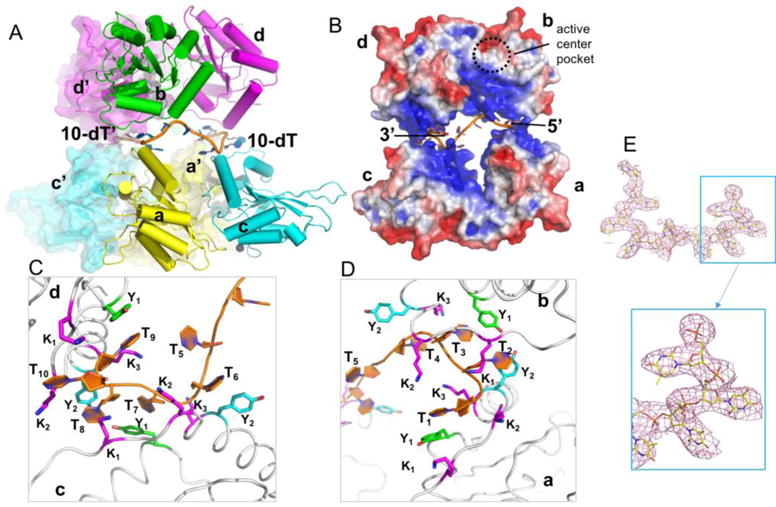
Crystal structure of A3F-CD2 in complex with a 10 nt poly-dT ssDNA. (A) The octameric crystallographic packing of A3F-CD2 in complex with two copies of 10 nt poly-dT ssDNA in one ASU. (B) Surface electrostatic potential of one 10 nt poly-dT ssDNA with four copies of A3F-CD2 molecules, colored according to calculated electrostatic potential of accessible surface area from −5 kT/e (red) to 5 kT/e (blue). The dashed circle represents the location of a proposed active center pocket. (C, D) Zoomed-in views of the ssDNA (orange)-A3F-CD2 interface. Y1, Y333 (green); Y2, Y359 (cyan); K1-3, K352, K355, K358 (magenta). (E) The electron density map for the entire 10 nt poly dT ssDNA (top) with zoomed-in view (bottom) from the co-crystal structure of A3F-CD2 bound to ssDNA, showing strong density for the DNA. The 2FoFc map was drawn at 1.0 sigma level.
Because one ASU contains eight molecules of A3F-CD2 and two 10 nt dT ssDNA, with each ssDNA binding to four monomers of A3F-CD2, we tried to further understand the oligomerization status of A3F-CD2. During purification, the cleaved hA3F-CD2 eluted as two major peaks on size exclusion chromatography (SEC) (Supplement Figure 1A), one of which (peak 1) was large oligomers in the void volume, while the other (peak 2) had an estimated molecular weight of approximately 29 kD. The estimate fell between the calculated molecular weight (MWt) of a monomer (21 kD) and a dimer (42 kD), but is closer to the monomeric form. Such elution observations suggest a possible transient interaction between two molecules at any given moment, where the mix of monomeric and dimeric states resulted in a peak position larger than expected for a monomer alone. We then used dynamic light scattering (DLS) to test the purified hA3F-CD2 that was crystallized, alone and in the presence of 10 dT ssDNA. The protein formed aggregates at 25°C, but behaved mostly as a dimer, with some larger oligomeric forms (see supplement Figure 1B, 1C) at 8°C, the lowest temperature the DLS instrument could be adjusted to. Upon adding the 10 dT ssDNA, the dimer disappeared, and tetramer and some larger oligomers were observed. This result suggests the multimerization of A3F-CD2 occurs upon binding to the 10 nt ssDNA.
To compare the roles of both the electrostatic and hydrophobic interactions between this mostly positively charged surface and ssDNA that were observed in the crystal structure, we generated three mutant constructs for testing the effects on ssDNA binding strength: one triple mutant K352A/K355A/K358A, and two individual mutants Y333A and Y359A. The initial efforts in purifying full length (FL) A3F wt and its mutant proteins yielded multimeric aggregated forms with nucleic acids contamination. To avoid complications of interpreting the nucleic acid binding data to the aggregated FL-A3F proteins with contaminated nucleic acids, A3F-CD2 construct and its mutants were used for the nucleic acid binding study. Also because it is known that A3F-CD2, similar to other APOBECs, has little binding to dsDNA (18,56), only ssDNA binding was tested in this study. To avoid potential kinetic effects due to substrate turnover from catalysis during binding assays using various ssDNAs, the catalytically inactive E251A version of A3F-CD2 (designated as WTi) was used to generate all three mutants for this DNA binding assay. The three mutant constructs were soluble and showed similar SEC elution profiles as the wild-type (Supplement Figure 1A). The binding strength of A3F-CD2 WTi and the three mutants on a 10 nt poly-dT ssDNA was tested using EMSA. The results showed that the triple lysine mutant K352A/K355A/K358A essentially abolished ssDNA binding completely (Figure 2, Table 2), indicating that the electrostatic interactions are critical for the interactions between A3F-CD2 and poly-dT ssDNA. The Y333A mutant also significantly impaired the poly-dT ssDNA binding (Figure 2, Table 2). On the other hand, the Y359A construct showed no impairment, and instead appeared to show a slight increase in the strength of poly-dT ssDNA binding (Figure 2, Table 2). The differences between the two tyrosine mutations could potentially be rationalized from the co-crystal structure in that each ssDNA interacts with the Y333 of all four bound subunits, but interacts with only two of the four Y359, suggesting that Y333 plays a more important role in binding the poly-dT ssDNA.
Figure 2.
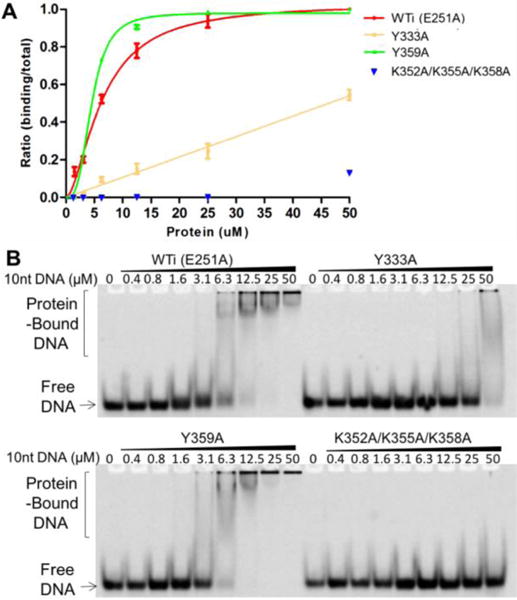
EMSA assay of A3F-CD2 and mutants binding to a 10 nt poly-dT ssDNA. The quantification and fitted curves are shown in (A) and one set of representative gels from three independent assays is shown in (B). Error bars were calculated in s.e.m.
Table 2.
The ssDNA/RNA binding of A3F-CD2 mutants.
| Kd | A3F-CD2 (E251A) | |||
|---|---|---|---|---|
| no extra mutation | Y333A | Y359A | K352A/K355A/K358A | |
| 10 nt poly-dT | 6.36±0.39 | N.D | 4.52±0.08 | N.D. |
| 30nt substrate ssDNA | 1.27±0.16 | 8.92±0.89 | 1.17±0.08 | N.D. |
| 50 nt RNA | 1.46±0.52 | 1.34±0.14 | 0.73±0.13 | N.D. |
Note: The binding affinity values (Kd) were estimated using native gel-shift assay (see Methods). N.D. indicates the value was difficult to be determine/measured due to the near background binding activity shown on the gel-shift assay.
The A3F-CD2 poly-dT interface is important for substrate binding and deamination
Considering the poly-dT ssDNA lacked a canonical deamination sequence motif, it was necessary to consider the role that substrate sequence has on the interaction with residues of the positively charged surface of A3F-CD2. Consistent with what we observed with a poly-dT substrate, a binding assay using a 30 nt substrate ssDNA containing a mixed DNA sequence and canonical TC motif for deamination showed a similar trend for the three mutants of A3F-CD2 (Figure 3A, B). Overall, binding of the 30 nt substrate with A3F-CD2 and all three mutants appeared to be stronger than with the 10 nt poly-dT (Figure 2, Figure 3A, B, Table 2), likely because of the length difference in the ssDNA allowing for stronger interactions. Both Y333A and K352A/K355A/K358A significantly impaired the substrate binding (Figure 3A, B, Table 2), whereas Y359A had slightly enhanced substrate binding. This result indicates that Y333 and K352/K355/K358 are involved in binding to the longer ssDNA consisting of a mixed sequence with a TC motif.
Figure 3.
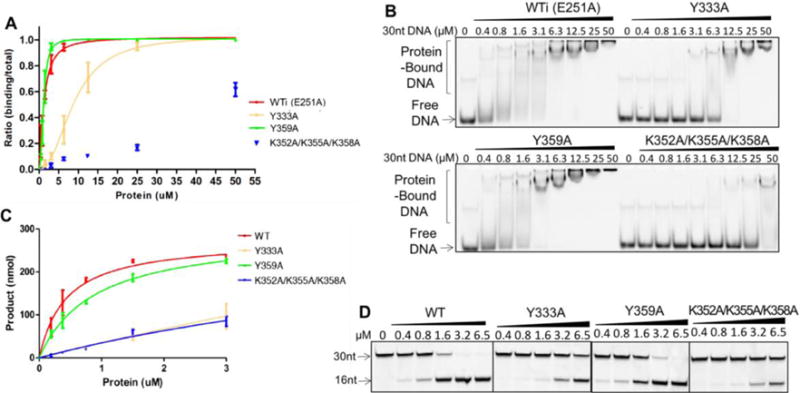
EMSA assay (A, B) and deamination assay (C, D) of A3F-CD2 and mutants with a 30 nt substrate ssDNA. For the DNA binding assay, A3F-CD2 catalytic E251A mutant (WTi) and the corresponding mutants were used to avoid complication of substrate turnover during DNA binding, but the wild-type catalytic E251 residue was restored in the corresponding mutants for the deamination assay. One set of representative gels from three independent assays for the DNA binding assay is shown in (B) and for the deaminase assay shown in (D). For the deaminase assay, the positions of the the 30 nt substrate ssDNA and the 16 nt deamination product ssDNA are indicated. Error bars were calculated in s.e.m. Deamination curves were fitted with the Michaelis-Menten model (see Methods).
In order to determine if the observed changes in ssDNA binding also affected deamination activity on the substrate DNA, we tested the same set of mutant A3F-CD2 constructs, this time with the wild-type catalytic E251 residue. The deamination activity was observed on the same 30 nt ssDNA substrate consisting of a mixed sequence with central TC motif. The deamination assay revealed that the catalytic activity of both the Y333A and K352A/K355A/K358A mutants were dramatically reduced by about 70% compared to WT A3F-CD2 (Figure 3C, D), suggesting that these residues are critical for the deamination activity of A3F-CD2, probably through mediating the interactions with the substrate ssDNA. Interestingly, although the Y359A mutant showed no reduction of ssDNA binding compared with wild type A3F-CD2 (Figure 3A, B), its catalytic activity was reduced (Figure 3C, D) (Table 2, Table 3).
Table 3.
Comparison of the Km values of A3F-CD2 WT and mutants.
| Km | WT A3F-CD2 | Y333A | Y359A | K352A/K355A/K358A |
|---|---|---|---|---|
| 30 nt substrate ssDNA | 0.43±0.01 | 24.22±87.07 | 0.88±0.13 | 8.99±8.10 |
Note: It should be noted that, since the product formed for two of the mutants (Y333A and K352A/K355A/K358A) could not reach saturation even when we used the maximum protein concentration obtained for these two constructs, the values obtained for these two constructs should be approximate.
A3F-CD2 binds ssDNA and RNA differently through the identified positively-charged surface
We next tested the RNA binding of A3F-CD2 WTi (containing the catalytic inactive E251A mutation) and the positively charged surface mutants by gel shift assays with a 50 nt RNA containing an unstructured, mixed sequence (Figure 4). The results showed that all the mutants had reduced RNA binding when compared to A3F-CD2 WTi (Figure 4A). In general, the amount of shifted RNA was significantly less than ssDNA, even though the length of tested RNA was longer than the tested ssDNA (containing 30 nt). For example, at a 50 μM A3F-CD2 protein concentration, almost 100% of the ssDNA was shifted with both the A3F-CD2 WTi and mutants Y333A and Y359A (Figure 3B), whereas only 50% of the RNA was shifted with the WTi (Figure 4B), and only ~30% of the RNA was shifted with mutants Y333A and Y359A. RNA binding by mutant K352A/K355A/K358A was not detectible under experimental conditions. Furthermore, for ssDNA binding, the Y359A appeared to be slightly stronger than WTi, and Y333A was significantly weaker (Figure 3A); whereas for RNA binding, both Y359A and Y333A were significantly weaker than A3F-CD2 WTi (Figure 4A). These results suggest that, even though these mutated residues of A3F-CD2 are involved in binding nucleic acids, their exact role in interacting with ssDNA and RNA appears to differ, and a more detailed understanding of their interaction mode would require a comparison between complex structures of A3F-CD2 binding to ssDNA and RNA molecules.
Figure 4.
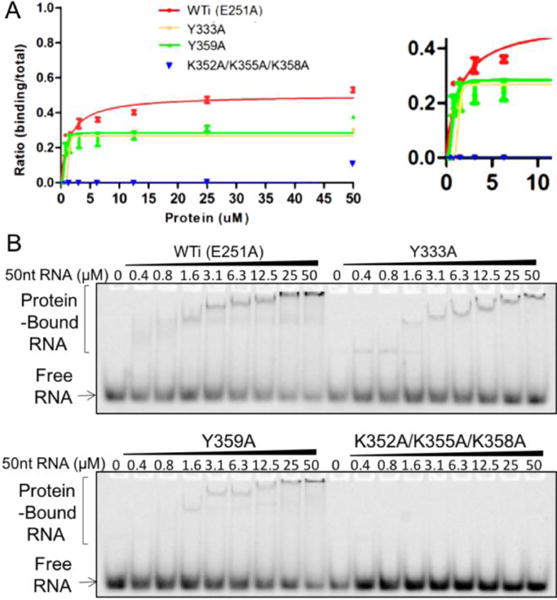
EMSA assay of A3F-CD2 and mutants binding to a 50 nt substrate RNA. The quantification and fitted curves are shown in (A) and one set of representative gels from three independent assays is shown in (B). Error bars were calculated in s.e.m.
Interestingly, when comparing the molecular surface charge features of A3F-CD2 with the available CD2 structures of A3G, another potent anti-HIV double domain APOBEC, it is obvious that the positively-charged surface identified for ssDNA binding in A3F-CD2 is actually negatively charged on A3G-CD2 (Figure 5A, 5B). Analysis of a sequence alignment between A3F-CD2 and A3G-CD2 showed that the equivalent residues to Y359 and the three positively charged residues K352, K355, and K358 in A3F-CD2 are negatively charged or neutral (E366, Q359, D362, D365) in A3G (Figure 5C), with only residue Y333 aligned between the two proteins.
Figure 5.
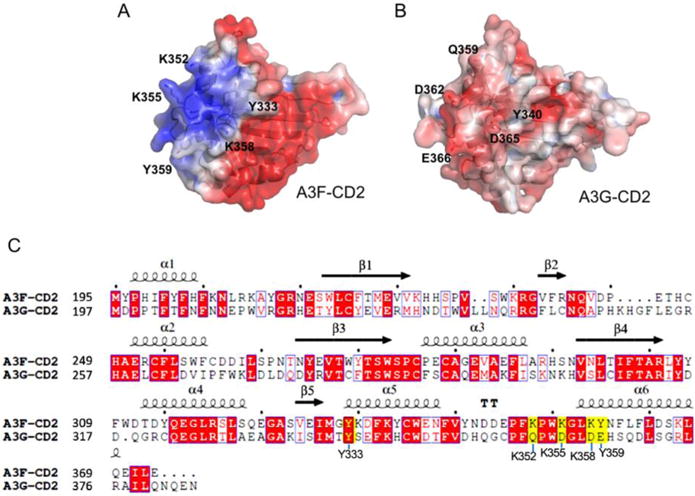
Sequence and structural comparison between A3F-CD2 and A3G-CD2. (A, B) Surface electrostatic potential of A3F-CD2 (A) and A3G-CD2 (B), colored according to calculated electrostatic potential of accessible surface area from −5 kT/e (red) to 5 kT/e (blue), as calculated by APBS (http://nbcr-222.ucsd.edu/pdb2pqr_2.1.1/). (C) Sequence alignments between A3F-CD2 and A3G-CD2. Depiction of secondary structural elements are shown above the sequences. Yellow highlighted residues are shown in the structure to interact with the poly-dT ssDNA and form an A3F-CD2 specific positively charged patch (A), which corresponds to a negatively charged patch on A3G-CD2 (B). The sequence alignment graphics were generated using ESPript.
An alternative inter-molecular zinc-coordination at the A3F-CD2 catalytic center
The typical catalytic center of APOBEC proteins contains a zinc atom coordinated by a histidine and two cysteines, which are essential for catalytic activity. All but one of the previously determined crystal structures of APOBEC proteins have so far had the typical zinc-coordinated active center conformation (Figure 6A) (53,54,56). Only one of the previously determined structures of various A3F-CD2 constructs was shown to be zinc-free, which was obtained at low pH (57). In the co-crystal structure of WT A3F-CD2 in complex with a poly-dT ssDNA, we found two A3F-CD2 molecules of a tetrameric unit had no Zn atom at the active center, and the other two A3F-CD2 molecules had a Zn atom that is located about 5 Å away from the canonical Zn position and with a different coordination conformation. The zinc atom is coordinated by three residues: H247 and H249 on loop 3, as well as residue H228 from loop 2 of an adjacent molecule (Figure 6B). Among the three histidine residues involved in this alternative coordination, only H249 belonged to the canonical active center residues. This suggests that A3F-CD2 has the potential to form alternative zinc coordination in solution beyond what is normally observed at the active Zn center. In order to test if this slightly off-positioned Zn atom has a role in regulating the catalytic activity by competing with the active-center Zn needed for deamination, we mutated H228 and H247 to abolish the alternative Zn-coordination. These mutants showed very similar deamination activity to the wild-type (Figure 6C). This result suggests that the alternative zinc-coordination conformation observed in the crystal structure has no critical role in regulating the catalytic activity in vitro. At this moment, it is not clear what this alternative Zn conformation means for the functions of A3F in vitro and in vivo. Combined with previous reports showing that A3F-CD2 can have Zn-free or Zn-coordination forms in different pHs (53,54,56,57), it appears that A3F-CD2 is unique in that its active center Zn may be less tightly held by the active center coordination bonds than those in other APOBEC members.
Figure 6.
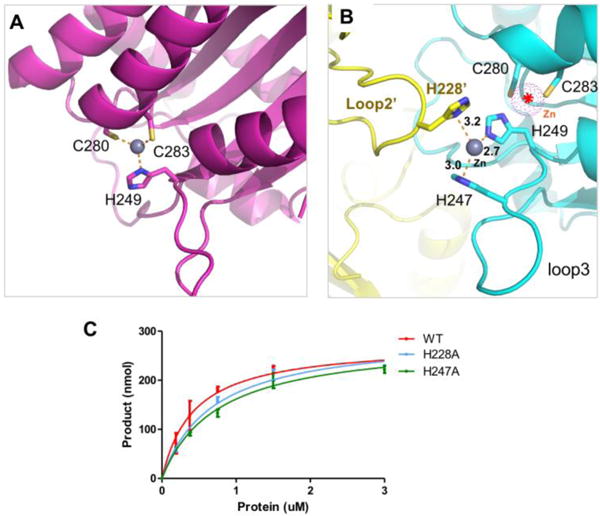
The alternative Zn-coordination in the A3F-CD2-poly-dT structure. (A) The classic intramolecular Zn-coordination within the active center of a wild-type A3F-CD2 structure (3WUS) (32). The Zn atom is drawn as a sphere (colored in grey), with three residues coordinating the Zn atom (H249, C280, C283) drawn in sticks. (B) The non-canonical Zn-coordination between two A3F-CD2 molecules is mediated by two histidines from loop 3 of one molecule and one histidine from loop 2 of another. The Zn atom is located about 5 Å away from the canonical Zn position (indicated by a dotted sphere in purple). The histidine (H249) that normally participates in the active center Zn coordination is also bound to this Zn atom. (C) The deamination assay of two mutants of the coordinating histidine residues that are observed to be involved in the inter-molecular Zn-coordination.
Discussion
A3F is a member of the APOBEC deaminase family of proteins that is capable of restricting HIV infection by targeting viral complementary ssDNA through a deaminase-dependent manner. The catalytic activity of these deaminases is not only determined by the catalytic center and surrounding loops, but also depends on how efficiently the enzyme binds to ssDNA substrates and allows the target sequence to access the active center. In this study, we reported the first co-crystal structure of the A3F-CD2 catalytic domain in complex with a 10 nt ssDNA consisting of a poly-dT sequence that reveals an unexpected ssDNA binding mode by this APOBEC protein. The structure revealed a positively-charged patch unique to A3F-CD2 that is involved with ssDNA binding. Mutating residues within this positively-charged interface significantly reduced the strength of ssDNA binding and dramatically impaired the deamination activity, suggesting that interactions mediated by this patch are indeed critically related to the catalytic activity.
The presented structure reveals that residues within this positively charged surface employ both electrostatic and hydrophobic interactions to bind ssDNA, and activity studies of mutants demonstrated that they are also related to substrate ssDNA binding and deamination activity. However, these two types of interactions appear to have different contributions. The electrostatic interactions are critical for both ssDNA binding and deamination, as both were dramatically reduced by mutating three lysine residues on the positively charged surface (Figure 3). However, the hydrophobic interactions revealed by the tyrosine mutations appear to disrupt deamination activity more profoundly than ssDNA binding activity (Figure 3). These results suggest that not all interactions with nucleic acids are equal in terms of their roles in affecting catalysis, i.e. some interactions with nucleic acids may be more “relevant” than others with respect to deamination activity. In this case, it is possible that the ssDNA substrates are bound to this positively-charged patch at different orientations non-specifically through electrostatic-interactions, but only some ssDNA substrates are “guided” by Y333 and Y359 through hydrophobic interactions to the specific orientation that is more favored for deamination.
It is noted that in the structure the ssDNA binding interface is not in proximity to the proposed active center pocket, and the orientation of the ssDNA does not point to the active center of the same A3F-CD2 molecule (Figure 1B). During purification, we also have observed that, under lower salt concentration (below 250 mM), A3F-CD2 tends to oligomerize and form large aggregates. Full length A3F is also shown to oligomerize into large molecular weight complexes both in vitro and in cells (31,63,64). The DLS results showed that A3F-CD2 forms tetramer or larger oligomers in the presence of 10 nt ssDNA, and that even larger oligomers are expected to form with longer ssDNA. Therefore, it is possible that this positively-charged patch may help capture and guide the ssDNA substrate for the deamination catalyzed by an adjacent A3F molecule in the A3F oligomeric complex.
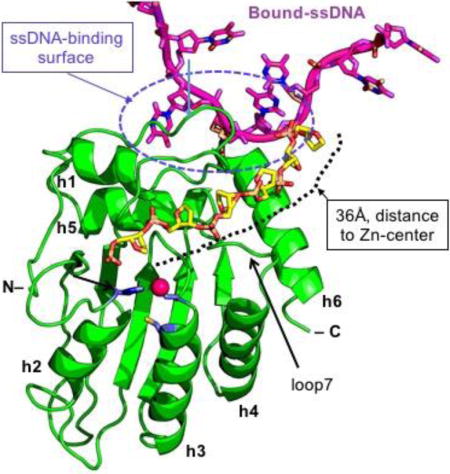
Even though the CD1 domain plays an important role in oligomerization of A3F and other double-domain APOBECs (32,33,37,45,49), the full-length A3F with a disrupted CD1 domain still forms high molecular weight complexes (65), suggesting that A3F-CD2 may also be involved in oligomerization. The crystal packing of the eight molecules within one ASU reported here, or those A3F-CD2 apo structures reported earlier (53,54,56,57) does not show an interface with significant buried areas for the formation of a stable dimer or multimer. One possible oligomerization interaction within the CD2 domain of A3F may be somewhat similar to what is observed for the dimer interactions of a primate A3G-CD1 (rA3G-CD2) involving residues around the C-terminal part of helix 6 (h6) (32). Similarly, a recent study of A3C suggested dimerization can occur in the presence of a S188I polymorphism around the C-terminal part of h6 (66). The equivalent residue 188 in A3F-CD2 is already isoleucine near the C-terminus of h6, away from the positively charged patch around the N-terminus of h6. Hence, A3F-CD2, which shares ~80% sequence identity with A3C, may also have a similar dimerization interface near the C-terminal end of h6 as in A3C and rA3G-CD1. A hypothetical model can be made for A3F CD2 dimerization based on the previously solved rA3G-CD1 dimer (32), which is categorized within the same Z2 hereditary domain class (Figure 7A). In this model, the angle between the two molecules is about 90°. When the structure of one A3F-CD2 molecule with bound ssDNA is superimposed onto the rA3G-CD1 dimer structure, the positively charged patch of A3F-CD2 binds the 5′ end of the DNA and guides the 3′ end towards the active center of another molecule (Figure 7B). In this model, the active-center loop 7 of the second molecule is right next to the bound DNA, consistent with a previous report that loop 7 of A3F-CD2 is involved in substrate binding (31).
Figure 7.
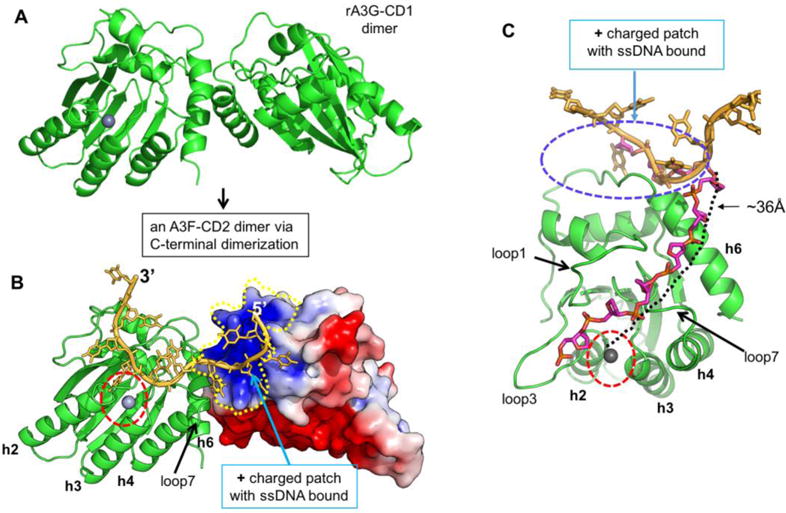
Hypothetical model of A3F-CD2 binding to a ssDNA at the positively charged patch as a way of catching a substrate DNA. (A) The dimer structure of rA3G-CD1 (PDB ID 5K82) (32) that dimerizes through the C-terminal end around h6. (B) A dimer model of A3F-CD2 based on the rA3G-CD1 dimer interface, with one copy of A3F-CD2 drawn as a charged surface and the bound 10 nt poly-dT shown in orange. The Zn-active center (indicated by red circle) of one molecule of the dimer is next to the ssDNA that has one end bound to the positively charged surface of the neighboring A3F-CD2 molecule. (C) A model showing how the bound ssDNA (colored in orange) interacting with the A3F-CD2 positively charged patch located away from the active center can take a different direction (the ssDNA backbone colored in purple) to extend to the active center of the same A3F-CD2. The path is approximately 36 Å in length, which would require about six nucleotides of ssDNA (with a conformation of roughly 6.3 Å per nt). The active center Zn is indicated by a red circle.
Alternatively, the bound ssDNA on the positively charged surface may be flexible enough to extend its target TC motif to the active center of the same A3F molecule, which is roughly 36 Å away from the bound ssDNA (or about the length of six nucleotides of ssDNA) (Figure 7C). In this model, the ssDNA could reach the active center by taking the shortest path over h6 and then passing between loop 1 and loop 7, around which there are several positively charged and hydrophobic residues that could interact with single stranded nucleic acids. A similar path over h6 and between loop 1 and loop 7 has been proposed previously for binding ssDNA (1,30). One common feature for both the aforementioned dimer model and the monomer model is that the ssDNA substrate is captured by a positively charged surface far away from the Zn-active center pocket, with the target C extended into the pocket. The surface electrostatics analysis of A3F-CD2 shows the area right around the Zn-active center pocket is surprisingly not highly positively charged (Supplement Figure 2). This feature is in contrary to that of A3A and A3G-CD1, which both are highly positively charged right around the Zn-active center pocket (Supplement Figure 2). This makes it a very interesting question as to whether that is the reason for the prior success in obtaining the co-crystal structures showing tightly bound ssDNA at the pocket of A3A (with a C) and A3G-CD1 (with a T) (32,60,61), but not bound at the pocket of A3F-CD2.
The proposed positively-charged patch provides a substrate binding site that is specific to A3F-CD2. This surface patch is in fact negatively charged in A3G-CD2. Interestingly, a recent study of mass spectrometry of A3G peptides cross-linked to nucleic acids mapped three spatially separated regions of A3G that cross-linked with DNA, one of which involved A3G-CD2 residues 345–374 around h6 (46). These A3G-CD2 residues partly overlap with the A3F-CD2 residues (333–358) located within its unique positively charged patch that were shown to bind substrate ssDNA. Without a structure of A3G-CD2 in complex with ssDNA, it is not clear how A3G would bind ssDNA through the three different regions, including the region around residues 345–373 in particular. Nevertheless, it does appear that A3G lacks a surface with positive charge in this region, which may explain the difference in strength of ssDNA binding and deamination activity observed for the individually expressed A3F-CD2 and A3G-CD2 domains. In A3F, it has been shown that the CD2 domain alone has easily detectable substrate binding and deamination, although the presence of its CD1 domain can enhance the strength of ssDNA binding as well as deamination activity by ~10 fold (31). On the contrary, in A3G, CD2 alone showed almost undetectable ssDNA binding, probably because it lacks this positively charged surface, and as such the CD1 domain for A3G appears to be much more critical for substrate binding and further enhancing deamination activity by ~1000 fold (30,38).
The structural and biochemical results reported herein may also imply that A3F can bind to RNA and ssDNA differently, as A3F-CD2 appears to bind ssDNA stronger than RNA (Figure 3, 4). Full-length A3F associates with RNA and has been shown to form higher-order ribonucleoprotein (RNP) complexes (67). Thus, most RNA binding may be mediated by the N-terminal non-catalytic CD1 domain, similar to A3G and A3B (49,68). In addition, we observed the lack of zinc atoms coordinated within the active center of the crystal structure, which is consistent with one previous report of an A3F-CD2 apo-structure that has no zinc present (57), further confirming that at least for A3F-CD2, the zinc coordination is not necessary in order to maintain a stable structure.
In summary, we solved a crystal structure of A3F-CD2 in complex with a 10 nt ssDNA consisting of a poly-dT sequence, and thus revealing the detailed interactions of ssDNA binding to an A3F-CD2-specific positively charged interface that is outside of the active center pocket, and demonstrating that key residues within this positively charged surface play a role not only in ssDNA binding but also in deamination catalysis. These findings provide novel insights about the interplay between APOBEC3F and DNA, and broaden understandings of the molecular mechanisms of the APOBEC cytidine deaminase family.
Methods
Protein expression and purification
A wild-type hA3F-CD2 construct (residues 190–373) was cloned into a pGEX-6p-1 vector with an N-terminal GST tag and PreScission cleavage site. Escherichia coli cells transformed with the plasmid were grown in LB media (37 °C) until the measured OD600 reached 0.25. Then, the media with cells were transferred into 16 °C shaker until OD600 reached 0.6, and 0.2 mM IPTG was added for overnight induction. The harvested bacteria cell pellets were resuspended in buffer I (25 mM Tris pH 8.0, 500 mM NaCl, 20 mM MgCl2, 1 mM DTT) with 0.1 mg ml−1 RNase A (Qiagen) and lysed via Microfluidizer. After centrifugation (4 °C, 12,000 rpm, 45 min), the supernatant of the cell lysates was incubated with GST resin for 2 hours, washed with buffer I, buffer II (50 mM Tris pH 8.0, 500 mM NaCl, 20 mM MgCl2, 0.5 mM TCEP) with a 500 mM, 1 M and 500 mM NaCl gradient and RNase A (0.05 mg ml−1), and eluted with buffer III (25 mM Glutathione, 50 mM Tris pH 8.8, 500 mM NaCl, 0.5 mM TCEP). GST-hA3F-CD2 was applied to a Superdex 200 gel filtration column (GE Healthcare) equilibrated with buffer II. Concentrated GST-hA3F-CD2 were then pooled, concentrated, and cleaved by PreScission protease in buffer. The cleaved hA3F-CD2 was then purified by SEC using a Superdex 75 gel filtration column at 4°C. The cleaved hA3F-CD2 showed two SEC elution peaks: a large peak of oligomers in the void volume, and second peak that eluted at an estimated molecular weight of 29 kDa. Protein in peak 2 was collected and concentrated for crystallization. A3F-CD2 mutants were purified by the same methods, which showed very similar SEC behavior as the wild-type.
Protein crystallization, data collection, structure determination and refinement
Purified hA3F-CD2 was concentrated to 5 mg ml−1 and incubated with 10 nt ssDNA poly-dT (protein: ssDNA=1:1.2) on ice for 1 hour for crystallization screening. Tiny rod-shape crystals were obtained by sitting drop vapor-diffusion method in 0.1 M potassium chloride, 0.005 M Magnesium Sulfate, 0.05 M HEPES PH=7.0, 15%(v/v) MPD. Diffraction data was collected from Advanced Photon Source 23-ID by 5 μm microbeam using the vector method. Data sets were indexed, integrated and scaled using the HKL2000 program package. The structure of hA3F-CD2 was determined by molecular replacement using molrep (CCP4 suite) with A3F-CD2 (PDB: 3WUS) as the template. After the first round of refinement, the poly-dT ssDNA was manually built in Coot based on the electron density. The complex structure was further refined by PHENIX and checked in Coot manually. The statistics for diffraction data and structural determination/refinement is shown in Table 1.
Deamination assay
A deamination assay was modified and expanded on from previous reports (69,70). Briefly, the purified wild type hA3F-CD2 and mutants were incubated with 5′-FAM labeled 30 nt ssDNA (5′ ATT TAT ATT ATT TAT TCA TAT TTA TAT TTA 3′) in a buffer consisting of 100 mM HEPES pH 7.2, 100 mM NaCl (10 μl reaction volume), and incubated at 37 °C for 2 h, followed by a brief inactivation at 90 °C for 5 min. Two units of uracil-DNA glycosylase was added to the reaction mixture, followed by incubation at 37 °C for 1 h. The reaction mixture was then mixed with 100 mM NaOH and heated at 90 °C for 10 min. The samples were then mixed with an equal volume of formamide and analyzed by 20% acrylamide urea-denaturing PAGE. Data from three independent assays were fit using a Michaelis-Menten model and the Km was calculated (Table 3). It should be noted that, since the product formation for two of the mutants (Y333A and K352A/K355A/K358A) could not reach saturation even when we used the maximum protein concentration obtained for these two constructs, the values obtained for these two constructs should be approximate.
Electrophoresis mobility shift assay (EMSA)
EMSA was completed as previously described(49). A3F-CD2 and mutants in combination with the inactivating mutation E251A were incubated with 5′-FAM labeled 10 nt ssDNA with a poly-dT sequence, a 30 nt ssDNA (5′ ATT TAT ATT ATT TAT TCA TAT TTA TAT TTA 3′) and a 50 nt RNA (AUU AUU AUU AUU UAU CCC UAU UUA UAU UUA UUG UUA UUG UUA UUG UUA UA) on ice for 10 min, in a buffer condition of 60 mM Tris pH 8.0, 100 mM NaCl (10 μl reaction volume). The reaction mixture was then mixed with 2 μl of 80% glycerol and analyzed by native PAGE. Un-shifted and shifted (including smears) complex on the gel was carefully quantified. Because of the formation of different oligomeric species when adding ssDNA/RNA to the purified A3F-CD2 in solution (especially the longer oligonucleotides), the DNA binding data from this EMSA assay were directly used to estimate Kd values for ssDNA/RNA binding. Because the DNA binding data shows some degree of cooperativity for the A3F-CD2 WTi and some of the mutants with different oligonucleotides, data from three independent assays were fitted by a non-linear one site-specific binding model with Hill slope model and Kd values were calculated (Table 2).
Dynamic light scattering measurement of hA3F-CD2 alone and with 10 dT ssDNA
Dynamic light scattering (DLS) was used to measure the molecular weight of hA3F-CD2 with or without 10 dT ssDNA. DLS measurements were performed using a DynaPro Titan (Wyatt Technology Corp.). The samples were centrifuged at 12,000g at 4°C for 10 minutes before making the measurements. The protein was not stable at room temperature, where the DLS is normally used. We then lowered the temperature of the sample chamber to 8°C (the lowest it can get to), which appeared to stabilize the protein. The data were collected with 30 acquisitions of 5 seconds each. The regularization analysis was used to analyze the data with the software DYNAMICS 7.1.7.
Data availability
Atomic coordinates and structure factors for hA3F-CD2 in complex with 10 poly-dT ssDNA have been deposited in the Protein Data Bank (PDB) with the accession codes 5W2M. All other data are available from the authors on reasonable request.
Supplementary Material
Highlights.
The first co-crystal structure of ssDNA binding to APOBEC outside the Zn-active center
Structure of wild-type CD2 domain of APOBEC3F (A3F) in complex with a well-ordered 10 nt ssDNA, which is the longest ordered ssDNA visible in a co-crystal structure with APOBEC.
A new nucleic acid binding surface of A3F-CD2 important for both ssDNA and RNA binding.
A new binding surface distal to active center for substrate binding and for catalytic activity of A3F-CD2
Acknowledgments
We thank BL-8.2.1 of Advanced Light Source at Berkeley, 19ID and 23ID of Advanced Photon Source at Argonne National Laboratory and the staffs there for assisting data collection, the core laboratory in the Center of Excellence in NanoBiophysics at University of Southern California (USC) for assisting biophysics characterization.
Funding: This work was supported by the National Institutes of Health grant R01GM087986 to X.S.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement. None declared.
References
- 1.Bransteitter R, Prochnow C, Chen XS. The current structural and functional understanding of APOBEC deaminases. Cell Mol Life Sci. 2009;66:3137–3147. doi: 10.1007/s00018-009-0070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salter JD, Bennett RP, Smith HC. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci. 2016;41:578–594. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitamura S, Ode H, Iwatani Y. Structural Features of Antiviral APOBEC3 Proteins are Linked to Their Functional Activities. Front Microbiol. 2011;2:258. doi: 10.3389/fmicb.2011.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov. 2015;5:704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imahashi M, Nakashima M, Iwatani Y. Antiviral Mechanism and Biochemical Basis of the Human APOBEC3 Family. Front Microbiol. 2012;3:250. doi: 10.3389/fmicb.2012.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niewiadomska AM, Yu XF. Host restriction of HIV-1 by APOBEC3 and viral evasion through Vif. Curr Top Microbiol Immunol. 2009;339:1–25. doi: 10.1007/978-3-642-02175-6_1. [DOI] [PubMed] [Google Scholar]
- 8.Hache G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS reviews. 2006;8:148–157. [PubMed] [Google Scholar]
- 9.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 10.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 13.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. Epub 2003 May 2028. [DOI] [PubMed] [Google Scholar]
- 14.Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Guo F, Cen S, Kleiman L. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology. 2007;365:92–100. doi: 10.1016/j.virol.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 16.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 17.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 18.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ –> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 19.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adolph MB, Webb J, Chelico L. Retroviral restriction factor APOBEC3G delays the initiation of DNA synthesis by HIV-1 reverse transcriptase. PloS one. 2013;8:e64196. doi: 10.1371/journal.pone.0064196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Sarkis PT, Luo K, Yu Y, Yu XF. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J Virol. 2005;79:9579–9587. doi: 10.1128/JVI.79.15.9579-9587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nature Medicine. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 25.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 26.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 27.Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Liu B, Liu X, Li Z, Yu XF, Zhang W. Identification of HIV-1 Vif Regions Required for CBF-beta Interaction and APOBEC3 Suppression. PloS one. 2014;9:e95738. doi: 10.1371/journal.pone.0095738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 30.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Q, Xiao X, Wolfe A, Chen XS. The in vitro Biochemical Characterization of an HIV-1 Restriction Factor APOBEC3F: Importance of Loop 7 on Both CD1 and CD2 for DNA Binding and Deamination. J Mol Biol. 2016;428:2661–2670. doi: 10.1016/j.jmb.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao X, Li SX, Yang H, Chen XS. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat Commun. 2016;7:12193. doi: 10.1038/ncomms12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chelico L, Prochnow C, Erie DA, Chen XS, Goodman MF. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J Biol Chem. 2010;285:16195–16205. doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 35.Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 37.Song C, Sutton L, Johnson ME, D’Aquila RT, Donahue JP. Signals in APOBEC3F N-terminal and C-terminal deaminase domains each contribute to encapsidation in HIV-1 virions and are both required for HIV-1 restriction. J Biol Chem. 2012;287:16965–16974. doi: 10.1074/jbc.M111.310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 41.Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, Tian C, Zhang W, Sarkis PT, Yu XF. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J Mol Biol. 2008;375:1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Li X, Ma J, Zhang L, Ma L, Mi Z, Zhou J, Guo F, Kleiman L, Cen S. Human APOBEC3F incorporation into human immunodeficiency virus type 1 particles. Virus research. 2014;191C:30–38. doi: 10.1016/j.virusres.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Polevoda B, Joseph R, Friedman AE, Bennett RP, Greiner R, De Zoysa T, Stewart RA, Smith HC. DNA mutagenic activity and capacity for HIV-1 restriction of the cytidine deaminase APOBEC3G depend on whether DNA or RNA binds to tyrosine 315. J Biol Chem. 2017;292:8642–8656. doi: 10.1074/jbc.M116.767889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donahue JP, Levinson RT, Sheehan JH, Sutton L, Taylor HE, Meiler J, D’Aquila RT, Song C. Genetic analysis of the localization of APOBEC3F to human immunodeficiency virus type 1 virion cores. J Virol. 2015;89:2415–2424. doi: 10.1128/JVI.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polevoda B, McDougall WM, Tun BN, Cheung M, Salter JD, Friedman AE, Smith HC. RNA binding to APOBEC3G induces the disassembly of functional deaminase complexes by displacing single-stranded DNA substrates. Nucleic Acids Res. 2015;43:9434–9445. doi: 10.1093/nar/gkv970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 48.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 49.Xiao X, Yang H, Arutiunian V, Fang Y, Besse G, Morimoto C, Zirkle B, Chen XS. Structural determinants of APOBEC3B non-catalytic domain for molecular assembly and catalytic regulation. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi K, Carpenter MA, Kurahashi K, Harris RS, Aihara H. Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain. J Biol Chem. 2015;290:28120–28130. doi: 10.1074/jbc.M115.679951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 53.Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, et al. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakashima M, Ode H, Kawamura T, Kitamura S, Naganawa Y, Awazu H, Tsuzuki S, Matsuoka K, Nemoto M, Hachiya A, et al. Structural Insights into HIV-1 Vif-APOBEC3F Interaction. J Virol. 2016;90:1034–1047. doi: 10.1128/JVI.02369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu X, Zhang T, Xu Z, Liu S, Zhao B, Lan W, Wang C, Ding J, Cao C. Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA. J Biol Chem. 2014 doi: 10.1074/jbc.M114.624262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siu KK, Sultana A, Azimi FC, Lee JE. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun. 2013;4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaban NM, Shi K, Li M, Aihara H, Harris RS. 1.92 Angstrom Zinc-Free APOBEC3F Catalytic Domain Crystal Structure. J Mol Biol. 2016;428:2307–2316. doi: 10.1016/j.jmb.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bohn MF, Shandilya SM, Silvas TV, Nalivaika EA, Kouno T, Kelch BA, Ryder SP, Kurt-Yilmaz N, Somasundaran M, Schiffer CA. The ssDNA Mutator APOBEC3A Is Regulated by Cooperative Dimerization. Structure. 2015;23:903–911. doi: 10.1016/j.str.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi K, Carpenter MA, Banerjee S, Shaban NM, Kurahashi K, Salamango DJ, McCann JL, Starrett GJ, Duffy JV, Demir O, et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat Struct Mol Biol. 2016 doi: 10.1038/nsmb.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kouno T, Silvas TV, Hilbert BJ, Shandilya SMD, Bohn MF, Kelch BA, Royer WE, Somasundaran M, Kurt Yilmaz N, Matsuo H, et al. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat Commun. 2017;8:15024. doi: 10.1038/ncomms15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitra M, Hercik K, Byeon IJ, Ahn J, Hill S, Hinchee-Rodriguez K, Singer D, Byeon CH, Charlton LM, Nam G, et al. Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res. 2014;42:1095–1110. doi: 10.1093/nar/gkt945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ara A, Love RP, Follack TB, Ahmed KA, Adolph MB, Chelico L. Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G. J Virol. 2017;91 doi: 10.1128/JVI.02230-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Dolan PT, Dang Y, Zheng YH. Biochemical differentiation of APOBEC3F and APOBEC3G proteins associated with HIV-1 life cycle. J Biol Chem. 2007;282:1585–1594. doi: 10.1074/jbc.M610150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adolph MB, Ara A, Feng Y, Wittkopp CJ, Emerman M, Fraser JS, Chelico L. Cytidine deaminase efficiency of the lentiviral viral restriction factor APOBEC3C correlates with dimerization. Nucleic Acids Res. 2017;45:3378–3394. doi: 10.1093/nar/gkx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gallois-Montbrun S, Holmes RK, Swanson CM, Fernandez-Ocana M, Byers HL, Ward MA, Malim MH. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J Virol. 2008;82:5636–5642. doi: 10.1128/JVI.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu Y, Ito F, Zhang G, Fernandez B, Yang H, Chen XS. DNA cytosine and methylcytosine deamination by APOBEC3B: enhancing methylcytosine deamination by engineering APOBEC3B. Biochem J. 2015;471:25–35. doi: 10.1042/BJ20150382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu J, Chen Q, Xiao X, Ito F, Wolfe A, Chen XS. Biochemical Characterization of APOBEC3H Variants: Implications for Their HIV-1 Restriction Activity and mC Modification. J Mol Biol. 2016;428:4626–4638. doi: 10.1016/j.jmb.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for hA3F-CD2 in complex with 10 poly-dT ssDNA have been deposited in the Protein Data Bank (PDB) with the accession codes 5W2M. All other data are available from the authors on reasonable request.


