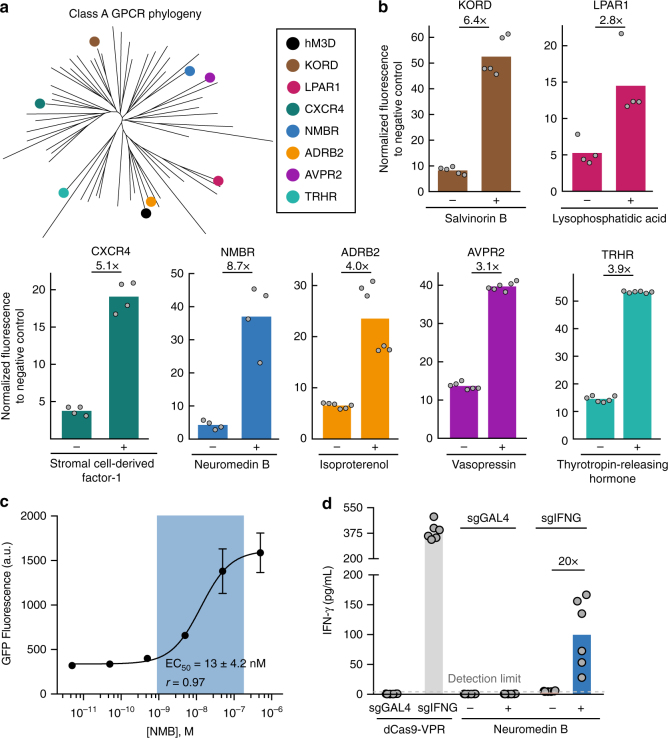
Fig. 4.
Expanding the ChaCha design to other synthetic and natural GPCRs. a The phylogenetic tree of Class A GPCRs. Synthetic (hM3D, KORD) and natural (LPAR, CXCR4, NMBR, ADRB2, AVPR, TRHR) GPCRs tested here are indicated. b The measured performance of diverse GPCRs with the ChaCha architecture for their respective ligands using HEK293T-GFP reporter cell line (see Supplementary Table 4 for ligand concentrations). The data were normalized to the free dCas9-VPR without a targeting sgRNA (for KORD, LPAR, CXCR4, NMBR) or with a non-targeting sgRNA (for ADRB2, AVPR, TRHR). The data represent two independent experiments with 2–3 technical replicates, and the bars represent the mean. For CXCR4, NMBR, and LPAR, we chose the best performing version characterized from a few variants as shown in Supplementary Fig. 9. See Methods section for the experimental procedure. c The dose–response curve of NMBR-CRISPR ChaCha in HEK293T cells after 1-day treatment of different CNO concentrations. EC50, the effective ligand concentration to achieve half-maximal GFP induction, and is shown as mean ± s.d. of three technical replicates. The replicate experimental data is shown in Supplementary Fig. 10. d Induction of endogenous IFN-γ by NMBR-CRISPR ChaCha in HEK293T cells after 2 days of 0.5 μM NMB treatment. sgGAL4, non-targeting sgRNA; sgIFNG, IFNG-targeting sgRNA.+/− indicates with or without Neuromedin B. The fold of activation displayed on top of bars compares+/− treatment conditions. The bars represent the mean, and the data represent two independent experiments with 3 technical replicates