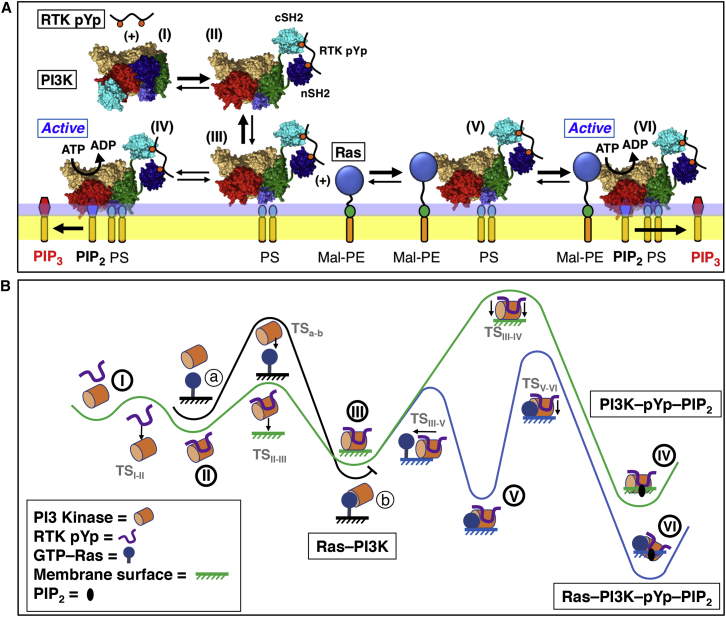
Figure 5.
Working model for the mechanism of synergistic PI3Kα activation by receptor-derived pYp and H-Ras. Shown are two different depictions of the same mechanistic model in which pYp peptide and H-Ras synergistically activate PI3Kα by enhancing its membrane recruitment. (A) Kinetic scheme depicting structural cartoons for each state in the hypothesized multi-step activation reaction, with dominant rates highlighted in bold. Activation begins with the binding of free PI3Kα in solution (I) to the receptor-derived pYp (RTK pYp), present in excess. One or both inhibitory SH2 domains bind pYp and dissociate from the PI3Kα catalytic domain, exposing its PIP2 binding and catalytic sites (9, 10). The resulting free pYp-activated PI3Kα complex (II) docks to the membrane, yielding an inactive pYp-PI3Kα complex transiently bound to the membrane surface in a shallow surface-bound state (III). This transient surface complex (III) rapidly dissociates and returns to solution (II) unless converted to a stable, membrane-bound pYp-PI3Kα complex by additional membrane interactions, such as deeper penetration into the bilayer (IV). The resulting stable, membrane-bound pYp-PI3Kα complex (IV) is fully active and displays the maximum specific kinase activity. Alternatively, if present, membrane-anchored H-Ras can rapidly bind to the transient surface complex (III) and catalyze its conversion to the stably bound, deeply penetrating, active Ras-pYp-PI3Kα complex (V). Such catalytic assistance explains the faster rate of formation of the Ras-bound stable, active complex (V) compared to Rasless stable, active complex (IV). Notably, the Ras-bound complex (V) exhibits two unexpected properties, leading to the hypothesis that Ras binding perturbs the optimal membrane docking geometry displayed by pYp-PI3Kα in the Rasless complex (IV). First, the rate of Ras-bound complex (V) dissociation from membrane is only slightly slower than that of Rasless complex (IV), indicating that the former complex does not possess the full kinetic stability expected for simple additivity of the binding free energies for pYp-PI3Kα binding to H-Ras plus pYp-PI3Kα binding to membrane. Second, the specific kinase activity of the Ras-bound complex (VI) is, surprisingly, significantly lower than that of the Rasless complex (IV). These unexpected features are both consistent with a Ras-bound state (VI) in which structural constraints prevent the optimal interactions of pYp-PI3Kα with Ras and the membrane simultaneously (see text). Further single-molecule studies will test this working model and determine the rate constants of individual steps. (B) Reaction coordinate depiction of the kinetic scheme shown in (A), using the same numbering scheme for intermediates (I) through (VI). Also shown is the unproductive path a-to-b for the absence of pYp, wherein PI3Kα is able to bind weakly to Ras but the lack of phospho-Tyr binding prevents release of the inhibitory SH2 domains bound to the catalytic domain. As a result, the SH2 domains continue to block PI3K membrane binding and kinase activation. To see this figure in color, go online.