ABSTRACT
Flavobacterium johnsoniae exhibits rapid gliding motility over surfaces. At least 20 genes are involved in this process. Seven of these, gldK, gldL, gldM, gldN, sprA, sprE, and sprT, encode proteins of the type IX protein secretion system (T9SS). The T9SS is required for surface localization of the motility adhesins SprB and RemA, and for secretion of the soluble chitinase ChiA. Here, we demonstrate that the gliding motility proteins GldA, GldB, GldD, GldF, GldH, GldI, and GldJ are also essential for secretion. Cells with mutations in the genes encoding any of these seven proteins had normal levels of gldK mRNA but dramatically reduced levels of the GldK protein, which may explain the secretion defects of the motility mutants. GldJ is necessary for stable accumulation of GldK, and each mutant lacked the GldJ protein. F. johnsoniae cells that produced truncated GldJ, lacking eight to 13 amino acids from the C terminus, accumulated GldK but were deficient in gliding motility. SprB was secreted by these cells but was not propelled along their surfaces. This C-terminal region of GldJ is thus required for gliding motility but not for secretion. The identification of mutants that are defective for motility but competent for secretion begins to untangle the F. johnsoniae gliding motility machinery from the T9SS.
IMPORTANCE Many members of the phylum Bacteroidetes secrete proteins using T9SSs. T9SSs appear to be confined to members of this phylum. Many of these bacteria also glide rapidly over surfaces using a motility machine that is also confined to the Bacteroidetes and appears to be intertwined with the T9SS. This study identifies F. johnsoniae proteins that are required for both T9SS function and gliding motility. It also provides an explanation for the link between secretion and gliding and identifies mutants with defects in motility but not secretion.
KEYWORDS: Flavobacterium, gliding motility, protein secretion, type IX secretion system
INTRODUCTION
Cells of Flavobacterium johnsoniae crawl rapidly over surfaces in a process known as gliding motility. This form of movement is common throughout the phylum Bacteroidetes, of which F. johnsoniae is a member (1). Gliding motility is also found in bacteria belonging to other phyla, but these appear to rely on motility machines that are unrelated to those of the Bacteroidetes (2–5). At least 20 Gld, Spr, and Rem proteins (Fig. 1) are involved in Flavobacterium gliding (6–11). Gliding involves the rapid movement of adhesins, such as SprB and RemA, along the cell surface (11–13). These are propelled along an apparently helical track by other components of the motility machinery that comprise the gliding “motor.” The exact nature of this motor is uncertain, but a proton gradient across the cytoplasmic membrane appears to be required for gliding, and recent results suggest that the motor may have a rotary component (14).
FIG 1.
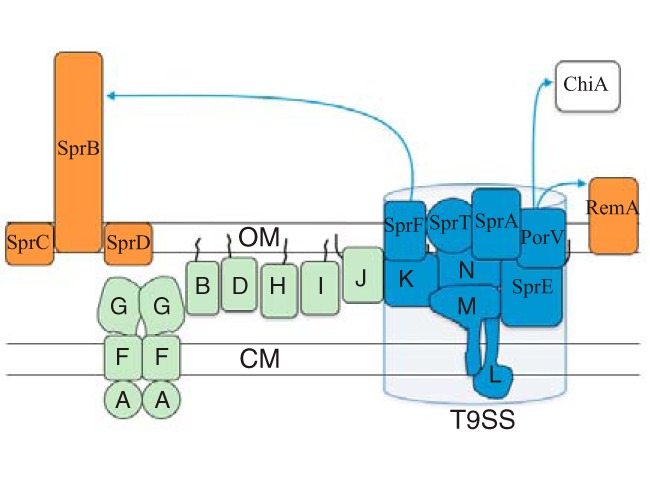
Proteins involved in F. johnsoniae gliding motility and protein secretion. SprB and RemA (orange) are motility adhesins that are propelled along the cell surface. SprC and SprD support SprB function. GldK, GldL, GldM, GldN, SprA, SprE, and SprT (blue) are components of the T9SS that are required for the secretion of SprB and RemA and for motility. The T9SS also secretes the chitinase ChiA (white), which is not involved in motility. PorV is needed for the secretion of RemA and ChiA but not SprB. SprF is needed for the secretion of SprB but not RemA or ChiA. Proteins in green (GldA, GldB, GldD, GldF, GldG, GldH, GldI, and GldJ) are also required for gliding motility. Black lines indicate lipid tails on lipoproteins. Proteins are not drawn to scale, stoichiometry of components is not known, and it is not known whether any of the lipoproteins localize to the cytoplasmic membrane (CM) instead of the outer membrane (OM), as shown.
Seven proteins (GldK, GldL, GldM, GldN, SprA, SprE, and SprT) are required for the secretion of SprB and RemA to the cell surface (7, 9, 10, 15). These proteins are thought to be the core components of the F. johnsoniae type IX secretion system (T9SS), which secretes these adhesins. The F. johnsoniae T9SS is also involved in the secretion of the soluble extracellular chitinase ChiA and many other proteins (16, 17). Other T9SS components that are not essential for its function include SprF (needed for secretion of SprB but not for secretion of RemA and ChiA) and PorV (needed for secretion of RemA and ChiA but not for secretion of SprB) (8, 11, 17). Mutations in the genes encoding any of the core components of the T9SS result in defects in gliding motility. These motility defects may be entirely the result of a failure to secrete the motility adhesins. Alternatively, some T9SS components may have functions in motility in addition to their roles in protein secretion.
T9SSs were originally discovered in the nonmotile periodontal pathogen Porphyromonas gingivalis and in F. johnsoniae (1, 15, 18). They are common in, but apparently confined to, the phylum Bacteroidetes (1, 19). Proteins secreted by the T9SS have N-terminal signal peptides that target them for export across the cytoplasmic membrane by the secretory (Sec) system, as well as conserved C-terminal domains (CTDs) that are required for secretion across the outer membrane by the T9SS (17–21). In most cases, the CTDs are thought to be removed after secretion by the activity of the protease PorU (22). Some proteins secreted by the T9SS, such as SprB, are attached to the cell surface and thus remain cell associated, whereas others, such as ChiA, are secreted in soluble form (10, 16). The mechanism by which SprB attaches to the cell surface is not yet known, but some P. gingivalis proteins secreted by the T9SS are covalently linked to outer membrane lipids (23, 24). In P. gingivalis, a complex composed of PorK, PorL, PorM, and PorN (orthologs of F. johnsoniae GldK, GldL, GldM, and GldN, respectively) has been characterized (15, 25, 26). This complex appears to span the cell envelope. It has been suggested that the cytoplasmic membrane-spanning proteins GldL and GldM (PorL and PorM, respectively, in P. gingivalis) may harvest the proton gradient to energize secretion (10, 26). The P. gingivalis orthologs of GldN and of the lipoprotein GldK appear to form ring-like structures on the inside of the outer membrane (25, 26). The outer membrane proteins SprA, SprE, and SprT (orthologs of the P. gingivalis T9SS proteins Sov, PorW, and PorT, respectively) are also required for secretion and may assist transit of the outer membrane, although the exact functions of these proteins are not known. Genome analyses revealed the cooccurrence of SprA with proteins carrying T9SS CTDs, suggesting the possibility that they interact (19).
P. gingivalis has orthologs of each of the F. johnsoniae T9SS genes, but it lacks orthologs of other F. johnsoniae genes involved in gliding, including gldA, gldD, gldF, gldG, gldH, gldI, gldJ, sprB, sprC, sprD, and remA. These F. johnsoniae genes were proposed to have roles in motility rather than in secretion, a suggestion that was supported by comparative analysis of the genomes of motile and nonmotile members of the phylum Bacteroidetes (1). However, indirect evidence suggests that mutations in some of these F. johnsoniae motility genes may cause T9SS defects. Cells with mutations in gldA, gldD, gldF, gldG, gldH, gldI, or gldJ are defective in chitin utilization and exhibit resistance to bacteriophages that are thought to use SprB and RemA as receptors (27–33). Here, we directly demonstrate that cells of these mutants exhibit T9SS defects and that they fail to accumulate the T9SS protein GldK, which may explain the secretion defects. Specific gldJ mutants that were deficient in gliding but competent for the secretion of SprB were identified, indicating that GldJ is required for gliding motility independent of its role in secretion. The F. johnsoniae gliding motility apparatus and T9SS appear to be intertwined. The identification of mutants with defects in motility but not secretion begins to untangle these two processes.
RESULTS
Mutations in gldA, gldB, gldD, gldF, gldH, gldI, or gldJ result in defects in secretion of ChiA and SprB.
GldK, GldL, GldM, GldN, SprA, SprE, and SprT are components of the T9SS and are required for the efficient secretion of SprB and ChiA (9–11, 15, 16, 34). The demonstrated motility defects of cells with mutations in the genes encoding these T9SS proteins (6, 7, 9, 15) may be explained by the absence of SprB and other motility adhesins on the cell surface. The roles of the other proteins that are required for gliding motility (GldA, GldB, GldD, GldF, GldG, GldH, GldI, and GldJ) are less clear. Cells with mutations in the genes encoding any of these proteins exhibit phenotypes, such as phage resistance and inability to digest chitin, that suggest T9SS defects (27–33). To directly test the effects of these genes on secretion, we examined wild-type and mutant cells for the presence of intracellular and secreted forms of ChiA and SprB. ChiA was found in the spent culture fluid of wild-type cells but not in the spent culture fluid of cells with mutations in gldA, gldB, gldD, gldH, gldI, and gldJ (Fig. 2). Instead, ChiA accumulated in the cells of the mutants, as has previously been demonstrated for T9SS mutants (10, 11, 16). In each case, complementation with the appropriate gld gene restored the secretion of ChiA. Similar results were obtained for the gldF mutant CJ787, which has a Tn4351 insertion in gldF that is polar on the downstream gene gldG (Fig. 2). CJ787 fails to produce both GldF and GldG (29). gldK mutant cells also failed to secrete ChiA, as expected given the demonstrated requirement of GldK for secretion (10). Previous results indicate that SprB is found on the surface of wild-type cells, whereas T9SS mutants accumulate SprB inside cells (9, 10, 13, 15, 34). This was confirmed here, since cells of the ΔgldK mutant failed to secrete SprB to the cell surface (Fig. 3). Cells with mutations in gldA, gldB, gldD, gldF, gldH, gldI, and gldJ behaved like the ΔgldK mutant. They produced SprB but failed to secrete it to the cell surface (Fig. 3). These results demonstrate that mutations in F. johnsoniae gldA, gldB, gldD, gldF, gldH, gldI, and gldJ result in T9SS defects. However, some other motility genes are not required for secretion. Cells with mutations in sprB, sprC, and sprD have severe but incomplete motility defects but retain the ability to digest chitin and to secrete chitinase (8).
FIG 2.
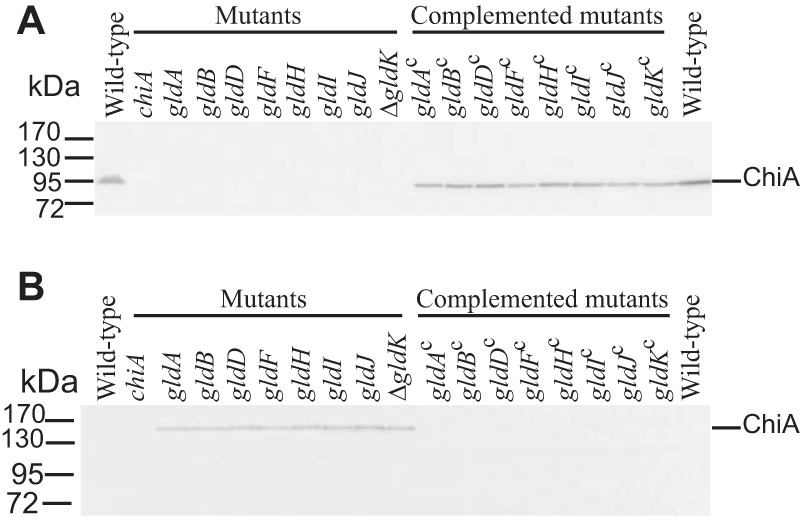
Mutations in gldA, gldB, gldD, gldF, gldH, gldI, and gldJ result in defects in secretion of the soluble extracellular chitinase ChiA. Cell-free spent medium (A) and cells (B) were examined for ChiA by SDS-PAGE, followed by Western blot analysis using antiserum against recombinant ChiA. ChiA is proteolytically processed during or after secretion, as previously reported (16), accounting for the smaller size in panel A. The cells analyzed were wild-type F. johnsoniae UW101, chiA mutant CJ1808, gldA mutant UW101-288, gldB mutant CJ569, gldD mutant CJ282, gldF mutant CJ787, gldH mutant CJ1043, gldI mutant UW102-41, gldJ mutant UW102-80, and ΔgldK mutant CJ2122. gldAc, gldBc, gldDc, gldFc, gldHc, gldIc, gldJc, and ΔgldKc are complemented versions of the mutants that carry pSA21, pDH223, pMM209, pMK314, pMM293, pMM291, pMM313, and pTB99, respectively. Samples loaded in panel B corresponded to 15 μg of protein per lane, and samples loaded in panel A corresponded to the volume of spent medium that contained 15 μg of cell protein before the cells were removed.
FIG 3.
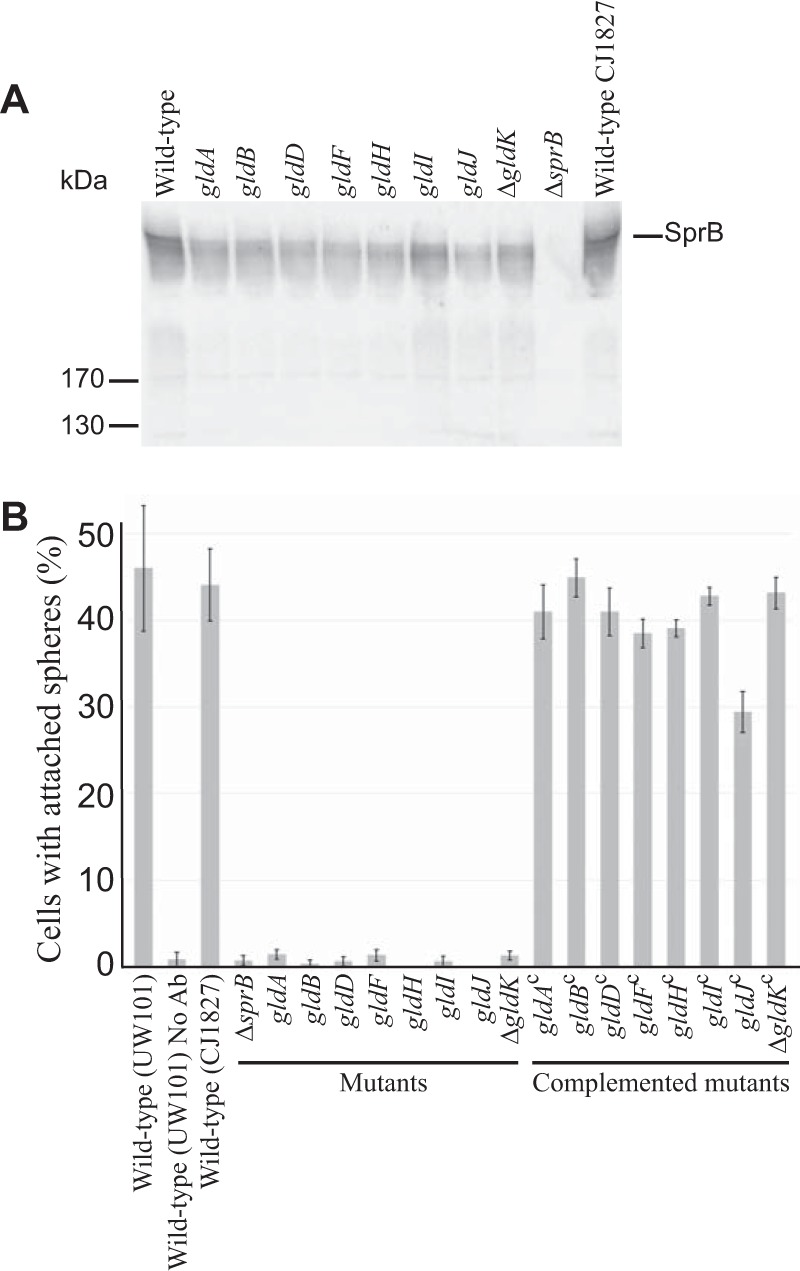
Cells with mutations in gldA, gldB, gldD, gldF, gldH, gldI, and gldJ produce the motility adhesin SprB but fail to secrete it to the cell surface. (A) Immunodetection of SprB in cells of wild-type or mutant F. johnsoniae strains. Whole cells were analyzed from cultures of wild-type F. johnsoniae UW101, streptomycin-resistant “wild-type” CJ1827, gldA mutant UW101-288, gldB mutant CJ569, gldD mutant CJ282, gldF mutant CJ787, gldH mutant CJ1043, gldI mutant UW102-41, gldJ mutant UW102-80, ΔgldK mutant CJ2122, and ΔsprB mutant CJ1922. Cell samples corresponded to 15 μg of protein per lane. Samples were separated by SDS-PAGE, and SprB was detected using antiserum against SprB. SprB is predicted to be 669 kDa after the removal of its N-terminal signal peptide, and it has previously been shown to migrate as a diffuse band (13). (B) Detection of SprB on the surface of wild-type or mutant cells. Anti-SprB antiserum and 0.5-μm-diameter protein G-coated polystyrene spheres were added to cells and examined using a phase-contrast microscope, as described in Materials and Methods. Images were recorded for 30 s, and 100 randomly selected cells were examined for the presence of spheres that remained attached to the cells during this time. Wild-type and mutant strains were as described in panel A. “No Ab” indicates no antisera were added to this sample. Complemented mutants (indicated by superscript “c”) carried the appropriate plasmids to express GldA (pSA21), GldB (pDH223), GldD (pMM209), GldF and GldG (pMK314), GldH (pMM293), GldI (pMM291), GldJ (pMM313), and GldK (pTB99). Error bars indicate standard deviations from three measurements.
Mutations in gldA, gldB, gldD, gldF, gldH, gldI, or gldJ result in dramatically reduced levels of the T9SS protein GldK.
gldK, gldL, gldM, and gldN comprise an operon in which each gene is required for T9SS function (10). Levels of GldK, GldL, GldM, and GldN were examined in wild-type and mutant cells. GldK was absent or present at greatly reduced levels in cells with mutations in gldA, gldB, gldD, gldF, gldH, gldI, or gldJ (Fig. 4). Since none of these genes reside near the gldKLMN operon, the absence of GldK was not the result of polarity. Further, the genes downstream of gldK were expressed, since GldL, GldM, and GldN were present in each of the seven mutants (Fig. 4). The level of gldK mRNA in wild-type and mutant cells was examined by quantitative reverse transcription-PCR (RT-PCR). The nearly identical cycle thresholds for the detection of gldK mRNA in wild-type cells and in cells with mutations in gldA, gldB, gldD, gldF, gldH, gldI, and gldJ indicate that similar levels of gldK mRNA were present in each (Fig. 5). A likely explanation for the absence of GldK protein in these mutants is the instability of GldK in the absence of the proteins encoded by these gld genes. gldA, gldB, gldD, gldF, gldH, and gldI are also each needed for the accumulation of stable GldJ protein, although they are not needed for the transcription of gldJ (28). A loss of GldJ in cells of the mutants may thus account for the absence of GldK protein. Although GldJ is required for the accumulation of stable GldK, GldK is not required for accumulation of the GldJ protein (6) (see Fig. S1 in the supplemental material).
FIG 4.
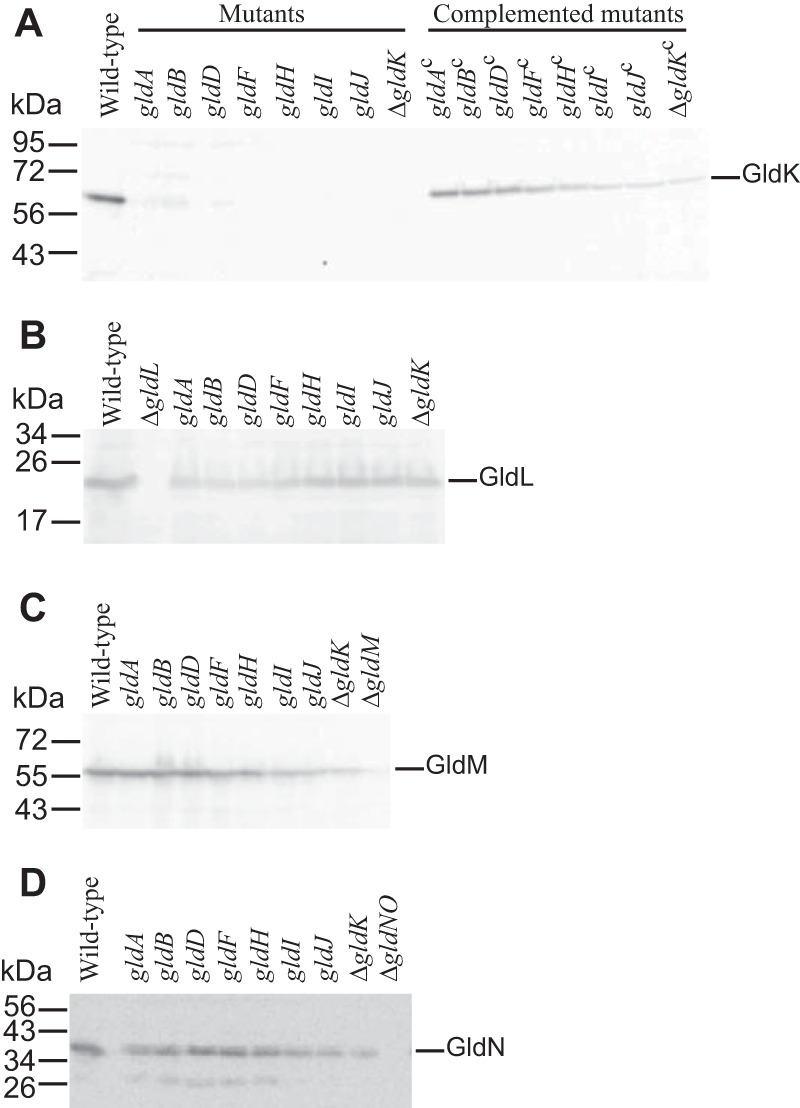
Immunodetection of GldK (A), GldL (B), GldM (C) and GldN (D) in cells of wild-type and mutant F. johnsoniae strains. Cell extracts (15 μg of protein) of wild-type (UW101), gldA mutant (CJ101-288), gldB mutant (CJ569), gldD mutant (CJ282), gldF mutant (CJ787), gldH mutant (CJ1043), gldI mutant (UW102-41), and gldJ mutant (UW102-80) were separated by SDS-PAGE, and GldK, GldL, GldM, and GldN were detected by Western blotting using the appropriate antisera (10, 34). Complemented mutants (indicated by superscript “c”) carried the plasmids indicated in Fig. 3. The ΔgldK mutant (CJ2122), ΔgldL mutant (CJ2157), ΔgldM mutant (CJ2262), and ΔgldNO mutant (CJ2090) were used as controls in panels A, B, C, and D, respectively.
FIG 5.
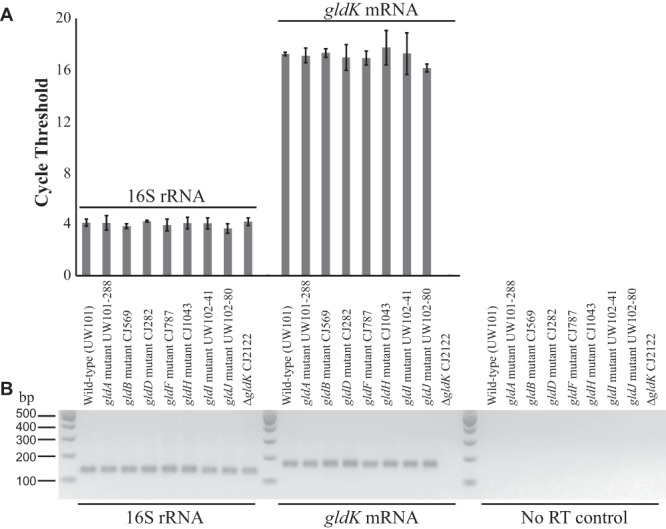
gldK mRNA levels in wild-type and mutant F. johnsoniae. cDNA was generated, and real-time quantitative PCR (qPCR) was performed as described in Materials and Methods. (A) RNA levels as indicated by the cycle number at which fluorescence exceeded the predetermined threshold for qPCR for 16S rRNA (using primers 1508 and 1509) and gldK mRNA (using primers 1506 and 1507), respectively. Error bars indicate 1 standard deviation of 6 runs (3 procedural replicates of 2 biological replicates). The ΔgldK CJ2122 sample probed for gldK mRNA never crossed the fluorescence threshold, indicating the absence of gldK mRNA. No-RT controls were samples prepared without reverse transcriptase that were amplified using primers 1508 and 1509. The predetermined thresholds were placed above the background fluorescence observed for these no-RT controls. (B) Agarose gel of pooled samples of the qPCR products. Strain labels apply to both panels.
Mutations in the T9SS genes sprA, sprE, and sprT did not dramatically affect GldJ or GldK levels.
Cells with mutations in sprA, sprE, and sprT also exhibit defects in gliding motility and protein secretion (9, 10, 15). To determine if these defects are the result of instability of GldJ and/or GldK, cells of the mutants were examined by Western blot analysis. Each of the mutants accumulated GldJ and GldK proteins, indicating that SprA, SprE, and SprT are not needed to stabilize GldJ and GldK (Fig. S1). SprA, SprE, and SprT are required for normal T9SS function (9, 10, 15), but the exact roles of these outer membrane proteins in secretion are not known.
Point mutations that alter the predicted active site of GldA lead to a loss of GldJ and GldK proteins and to defects in secretion.
Most of the Gld proteins are not similar to proteins of known function, complicating predictions of exact biochemical activities. GldA, however, is an exception. GldA is predicted to be the ATP-binding component of an ATP-binding cassette (ABC) transporter (27). The membrane proteins GldF and GldG are also components of this putative transporter (29). Disruption of F. johnsoniae gldA, gldF, or gldG results in a complete loss of motility (27, 29). The results presented above suggest that the proteins encoded by these genes are also needed for T9SS-mediated secretion.
F. johnsoniae strains with spontaneous and chemically induced point mutations in gldA were examined to determine whether GldA has a central role in motility, in protein secretion, or both. Two nonmotile mutants (UW102-9 and UW102-168) that had point mutations in gldA produced GldA protein, as detected by Western blot analyses (Fig. 6). Sequence analyses revealed that UW102-9 has a single nucleotide change (G118A numbered from the “A” of the gldA start codon), resulting in a G40R amino acid substitution in the predicted ATP-binding site of GldA, whereas UW102-168 has a C409T nucleotide change resulting in an R137C amino acid substitution in the ABC transporter signature sequence LSKGYRQR (site of change underlined). GldJ and GldK levels were dramatically reduced in both mutants, and complementation with wild-type gldA on pSA21 restored GldJ and GldK to wild-type levels (Fig. 6). These results suggest that the stable accumulation of GldJ and GldK requires not only the GldA protein but also GldA function. We do not know the function of GldA, but it presumably binds to and hydrolyzes ATP, and mutations predicted to disrupt this activity resulted in a loss of GldJ and GldK proteins.
FIG 6.
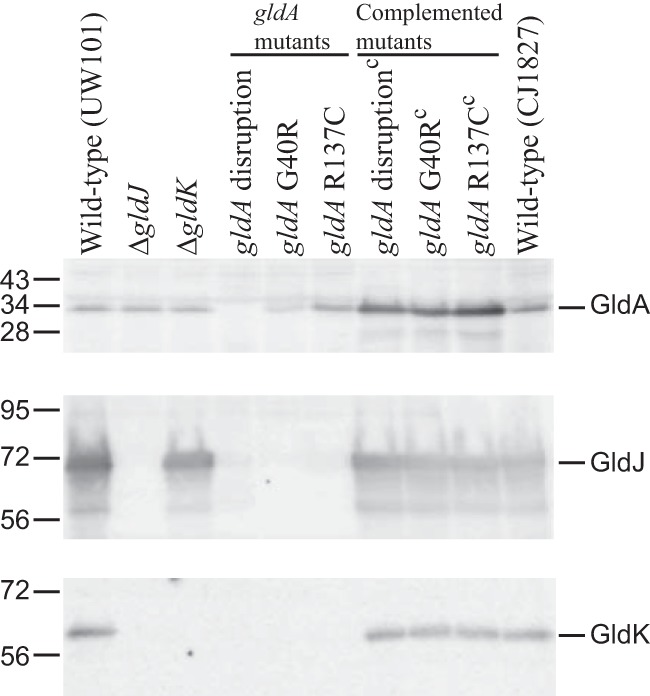
Immunodetection of GldA, GldJ, and GldK in wild-type cells and in cells of strains with mutations in gldA, gldJ, and gldK. UW101 is the F. johnsoniae type strain. CJ1827 is a streptomycin-resistant rpsl mutant of strain UW101 and is wild type for motility and secretion. The ΔgldJ and ΔgldK mutants are CJ2360 and CJ2122, respectively. “gldA disruption” refers to mutant CJ101-288. “gldA G40R” and “gldA R137C” refer to mutants UW102-9 and UW102-168, respectively, which produce GldA with the indicated amino acid changes. Strains listed with superscript “c” denote complementation with pSA21, which carries wild-type gldA.
GldK accumulates in cells expressing C-terminal truncated versions of GldJ.
If GldJ interacts with and stabilizes GldK, it is possible that some GldJ fragments would be sufficient to stabilize GldK, resulting in T9SS function, but would be insufficient to support motility, thus separating gliding motility from secretion. To identify such fragments, strains producing truncated forms of GldJ were generated. Plasmids encoding truncated forms of GldJ were introduced into the gldJ-null mutant UW102-55 (28). pMM317, pAB31, and pMM318 encode the first 337 (GldJ337), 542 (GldJ542), and 547 (GldJ547) amino acids of GldJ, whereas pMM313 encodes full-length GldJ (561 amino acids). These strains with plasmid-encoded truncated versions of GldJ were examined for the presence of GldJ and GldK proteins. Immunoblot analysis showed that GldJ and GldK were absent in cells of the gldJ mutant UW102-55 (Fig. 7A), which has a frameshift mutation (G inserted after position 88 numbered from the A of the gldJ start codon) (28). Cells of UW102-55 carrying pMM317 (which produces GldJ337) also accumulated no detectable GldJ and GldK proteins. In contrast, cells of UW102-55 carrying pAB31 accumulated a small amount of truncated GldJ542 but failed to accumulate GldK, and cells of UW102-55 carrying pMM318 accumulated GldJ547 and a small amount of GldK (Fig. 7A). Complementation of UW102-55 with pMM313 (expresses full-length GldJ561) resulted in large amounts of GldJ and wild-type levels of GldK.
FIG 7.
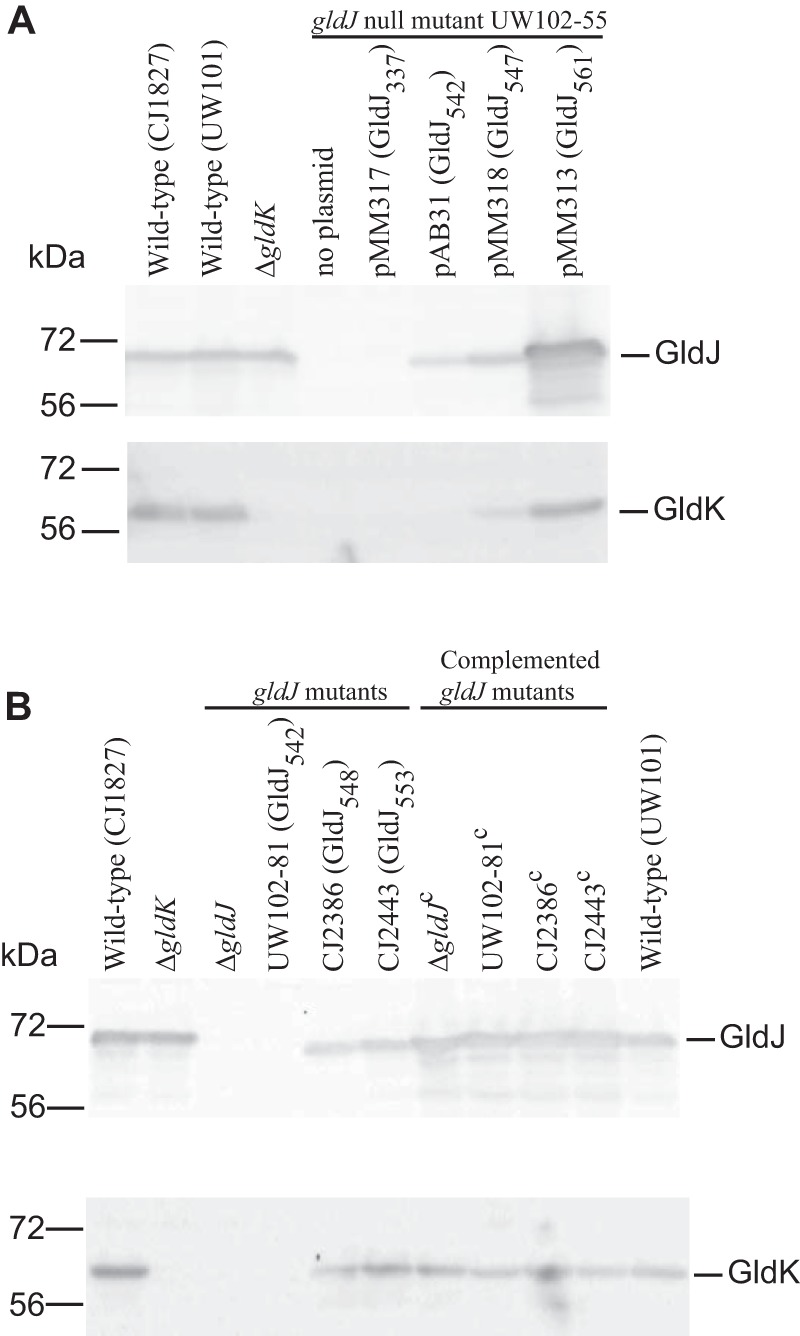
Immunodetection of GldJ and GldK in cells that produce truncated forms of GldJ. (A) Truncated forms of GldJ expressed from plasmids. Cells of wild-type, ΔgldK mutant, gldJ-null mutant UW102-55 (expresses the first 29 amino acids [aa] of GldJ), and UW102-55 carrying pMM317 that expresses the first 337 aa of GldJ (GldJ337), pAB31 that expresses the first 542 aa of GldJ (GldJ542), pMM318 that expresses the first 547 aa of GldJ (GldJ547), and pMM313 that expresses full-length GldJ (GldJ561) were examined for GldJ and GldK protein by Western blot analyses. (B) Truncated forms of GldJ expressed from the chromosome. Cells of the wild type, ΔgldK mutant, ΔgldJ mutant, and of chromosomal gldJ mutants that express the first 542, 548, and 553 aa of GldJ were examined for GldJ and GldK proteins by Western blot analyses. Strains listed with superscript “c” denote complementation with pMM313, which carries wild-type gldJ. For both panels, samples (15 μg of protein per lane) were separated by SDS-PAGE, and GldJ and GldK were detected using the appropriate antisera (10, 28).
The plasmid expression experiments described above suggest that a short region near the C terminus of GldJ may be dispensable for the accumulation of GldK. To explore this region in greater detail and to avoid issues of overexpression of GldJ from plasmid, we identified and constructed strains with truncated versions of gldJ on the chromosome. UW102-81 has a point mutation (A to T at position 1627, numbered from the A of the gldJ start codon) that results in a premature stop codon and termination of GldJ after amino acid 542 (Fig. 8) (28). CJ2386 and CJ2443 have in-frame deletions that terminate GldJ after amino acids 548 and 553, respectively. UW102-81 accumulated no detectable GldJ542, suggesting that the C-terminal 19 amino acids of GldJ may be required for its stability (Fig. 7B). In contrast, CJ2386 and CJ2443 produced GldJ548 and GldJ553, respectively. Each strain was also examined for levels of the GldK protein. As expected, UW102-81, which accumulated no GldJ, had no detectable GldK (Fig. 7B). In contrast, CJ2386 (produces GldJ548) and CJ2443 (produces GldJ553) each accumulated GldK protein.
FIG 8.
Alignment of C-terminal regions of GldJ proteins. GldJ sequences from 22 members of the phylum Bacteroidetes were aligned using MUSCLE. Dark shading indicates identical amino acids, and light shading indicates similar amino acids. Numbers at the right correspond to the length of each wild-type GldJ protein, including signal peptides. Arrows indicate the C-terminal amino acids of mutant F. johnsoniae GldJ proteins truncated at 542, 547, 548, and 553 aa. Sequences were organized with those most similar to F. johnsoniae GldJ at the top and those least similar at the bottom, with class of organisms within the phylum Bacteroidetes indicated on the left. See Fig. S2 for alignment of full-length GldJ sequences.
The C-terminal 13 amino acids of GldJ are required for motility but not for T9SS-mediated secretion of ChiA and SprB.
The ability of some truncated versions of GldJ to support accumulation of the T9SS protein GldK suggested that secretion might be functional in strains expressing these truncated forms of GldJ. Cells expressing GldJ542 failed to utilize chitin and failed to secrete ChiA into the culture fluid (Fig. 9A and B). Instead, these cells accumulated ChiA in the cells (Fig. 9C). This secretion defect was not surprising, since, as mentioned above, cells expressing GldJ542 failed to accumulate the T9SS protein GldK. In contrast, cells expressing GldJ547, GldJ548, and GldJ553 digested chitin and secreted ChiA, although cells expressing GldJ547 and GldJ548 did not accumulate as much extracellular ChiA as did wild-type cells. Similar results were observed for secretion of the motility adhesin SprB, except that cells expressing GldJ548 appeared to secrete SprB as well as did cells expressing full-length GldJ. CJ2360 (ΔgldJ) produced SprB (Fig. 10A) but failed to secrete it to the cell surface, as determined by a lack of attachment of anti-SprB-coated polystyrene spheres to these cells (Fig. 10B). In contrast, CJ2386 (expresses GldJ548) and CJ2443 (expresses GldJ553) secreted SprB, indicating the presence of functional T9SSs in these strains (Fig. 10B). The experiments described above used limiting amounts of anti-SprB to avoid large aggregates of cells and polyvalent spheres. In order to more accurately determine what percentage of cells harbored SprB on the cell surface, we examined cells by immunofluorescence microscopy using anti-SprB and the F(ab′) fragment of goat anti-rabbit IgG conjugated to Alexa Fluor 488 (Fig. 11 and S3). The results indicate that nearly all wild-type cells carried SprB on their surfaces, whereas cells of the T9SS ΔgldK mutant and of the ΔgldJ mutant that fails to accumulate GldK protein lacked surface-exposed SprB. In contrast, nearly all cells of the gldJ truncation mutants expressing GldJ548 and GldJ553 had cell surface SprB. The results of flow cytometry analyses (Fig. S4) confirm the presence of SprB on over 90% of the cells of the gldJ truncation mutants expressing GldJ548 and GldJ553. They also indicate that the levels of SprB on the surface of cells of these two gldJ truncation mutants and of the wild type were similar.
FIG 9.
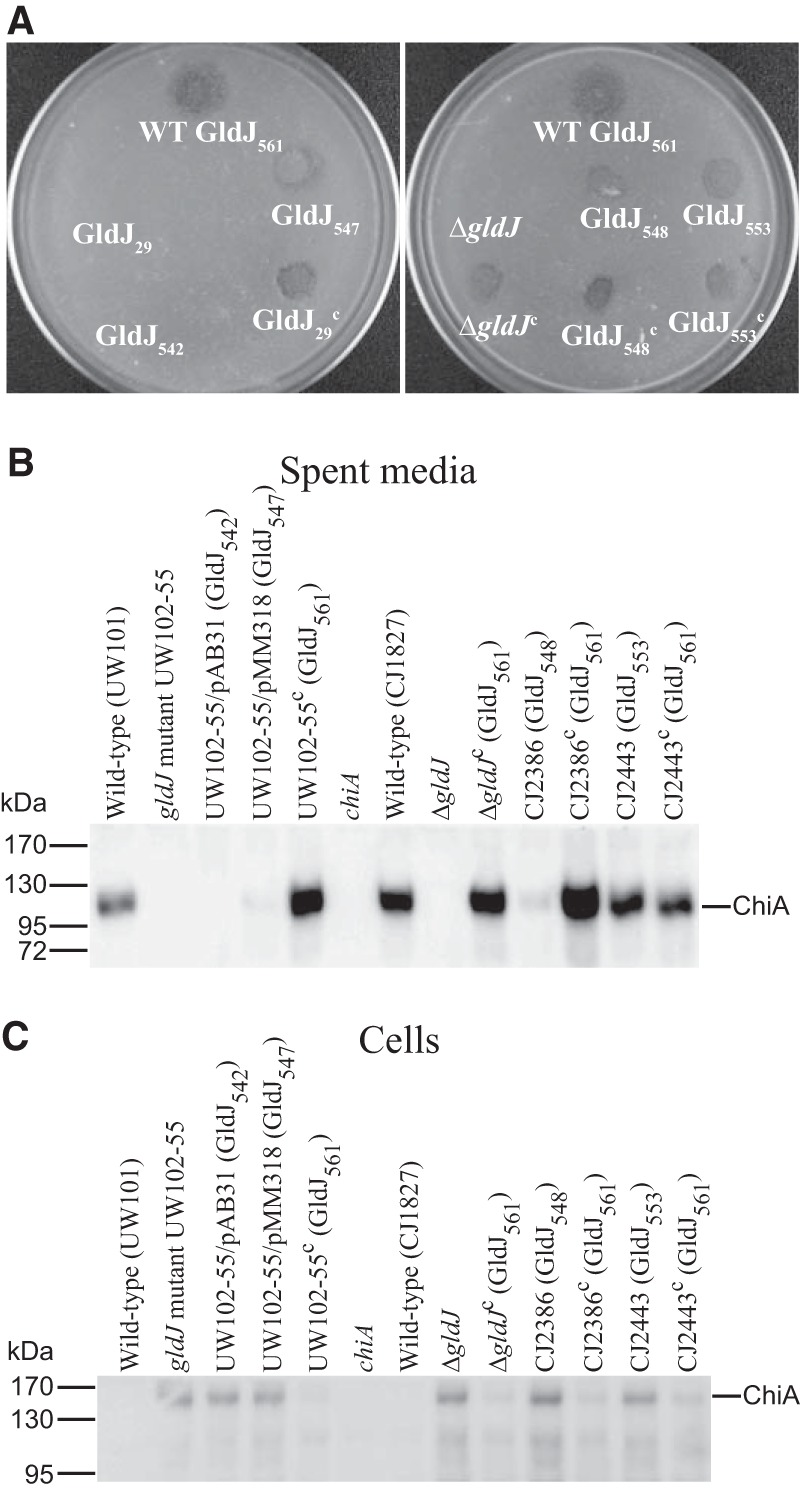
Effect of truncated GldJ on chitin utilization and secretion of ChiA. (A) Chitin utilization. Approximately 106 cells of wild-type or mutant F. johnsoniae were spotted on MYA-chitin medium and incubated at 25°C for 48 h. The plate on the left shows wild-type F. johnsoniae (WT; top), which expresses full-length GldJ (GldJ561); the gldJ-null mutant UW102-55, which expresses the first 29 aa of GldJ (GldJ29); UW102-55 carrying pAB31, which expresses the first 542 aa of GldJ (GldJ542); UW102-55 carrying pMM318, which expresses the first 547 aa of GldJ (GldJ547); and UW102-55 complemented with pMM313, which expresses full-length GldJ (GldJ29c). The plate on the right shows wild-type F. johnsoniae (expresses GldJ561), the ΔgldJ mutant, and the chromosomal gldJ mutants CJ2386 and CJ2443 that express the first 548 (GldJ548) and 553 (GldJ553) aa of GldJ, respectively. ΔgldJc, GldJ548c, and GldJ553c denote the indicated mutants complemented with pMM313, which carries wild-type gldJ. (B and C) Secretion of ChiA. Cell-free spent media (B) and cells (C) were examined for ChiA by SDS-PAGE, followed by Western blotting using antiserum against ChiA. Strains that were wild type for gldJ included UW101, the chiA disruption mutant CJ1808 (chiA), and the rpsl mutant of UW101, CJ1827. Plasmids pAB31 that expresses GldJ542, pMM318 that expresses GldJ547, and pMM313 that expresses full-length GldJ (GldJ561) were expressed in the gldJ-null mutant UW102-55. Plasmids encoding truncated GldJ are indicated by “/pAB31 (GldJ542)” and “/pMM318 (GldJ547).” CJ2386 and CJ2443 are chromosomal gldJ mutants that express the first 548 and 553 aa of GldJ, respectively. Samples loaded in panel C correspond to 15 μg of protein per lane, and samples loaded in panel B correspond to the volume of spent medium that contained 15 μg of cell protein before the cells were removed.
FIG 10.
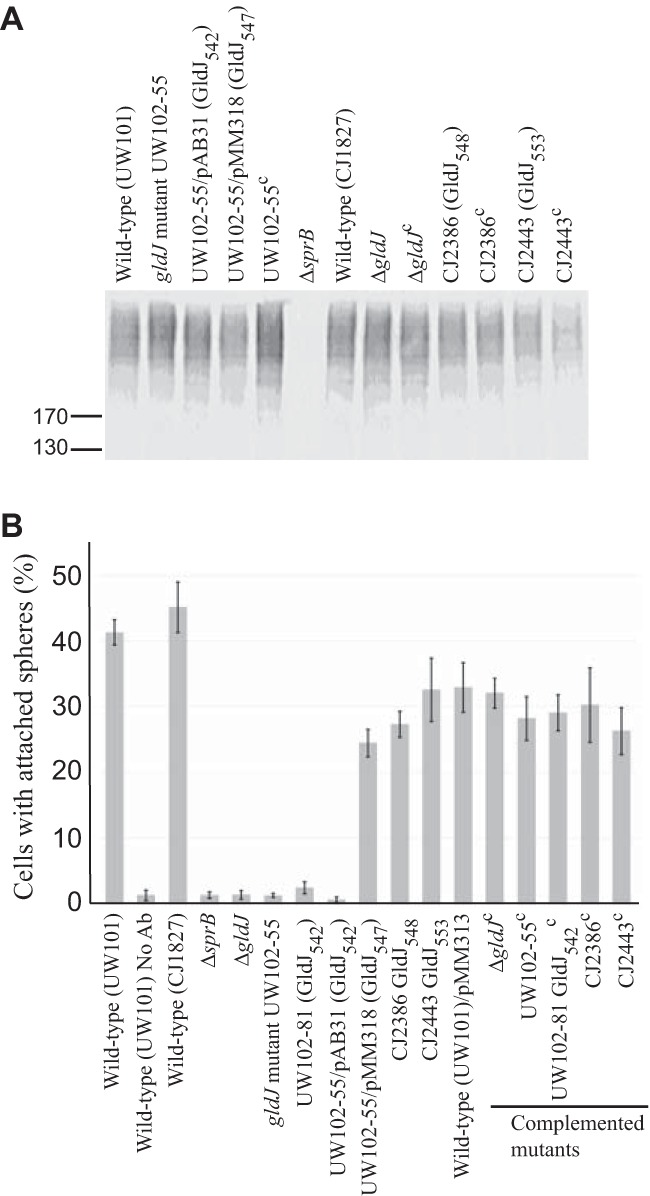
Secretion of SprB by cells with truncated GldJ. (A) Cell-associated SprB levels were examined by Western blotting. Cells of wild-type strains (UW101 and CJ1827), of gldJ-null mutants (UW102-55 and ΔgldJ mutant), and of strains expressing truncated forms of GldJ (indicated by “GldJ” followed by length in amino acids in subscript) were lysed in SDS-PAGE loading buffer, and proteins (15 μg per lane) were separated by SDS-PAGE and detected using antisera against SprB. Superscript “c” indicates complementation with pMM313 that carries wild-type gldJ (applies to panels A and B). CJ1922 (ΔsprB) was used as a negative control. (B) SprB on the cell surface was detected by adding anti-SprB antiserum and 0.5-μm-diameter protein G-coated polystyrene spheres to the cells. One hundred randomly selected cells were examined for the presence of spheres that remained attached to the cells during 30 s of microscopic observation. Error bars indicate standard deviations from three measurements. “No Ab” indicates no antisera added to this sample.
FIG 11.

Detection of surface-localized SprB by immunofluorescence microscopy. Cells of wild-type and mutant F. johnsoniae were exposed to anti-SprB antiserum followed by F(ab′) fragment of goat anti-rabbit IgG conjugated to Alexa Fluor 488. Cells of mutants CJ2386 and CJ2443 produce truncated versions of GldJ (548 and 553 aa, respectively). Images were recorded by immunofluorescence microscopy and by differential interference contrast (DIC) microscopy and then overlaid. The same exposure time was used for all fluorescence images. See Fig. S3 for original DIC and fluorescence images before merging. “No α-SprB” indicates no primary antiserum added to this sample. The scale bar indicates 10 μm and applies to all images.
The motility of strains expressing truncated versions of GldJ was also examined. Strains expressing GldJ542, GldJ548, and GldJ553 all produced nonspreading colonies (Fig. 12), indicating motility defects. Examination of cells in tunnel slides revealed dramatic defects in gliding for each of these strains (Fig. 13 and Movies S1 to S3). Cells expressing GldJ542 and GldJ548 were completely nonmotile, whereas cells expressing GldJ553 exhibited limited but detectable cell movement. Cells expressing GldJ542 failed to even bind to the glass slide, presumably as a result of a defect in secretion of cell surface adhesins, whereas cells expressing GldJ548 attached to glass but failed to glide. The introduction of pMM313 carrying wild-type gldJ restored gliding motility to each of the mutants. Although the complemented cells exhibited obvious movement on glass (Fig. 13 and Movies S1 to S3), they formed colonies that spread less well than those of the wild type, indicating a partial motility defect (Fig. 12, right side). This appears to be a result of overexpression of wild-type GldJ, since, as previously reported (28) and as confirmed here, the expression of gldJ from pMM313 (copy number, approximately 10) in wild-type cells also results in decreased spreading (Fig. 12).
FIG 12.
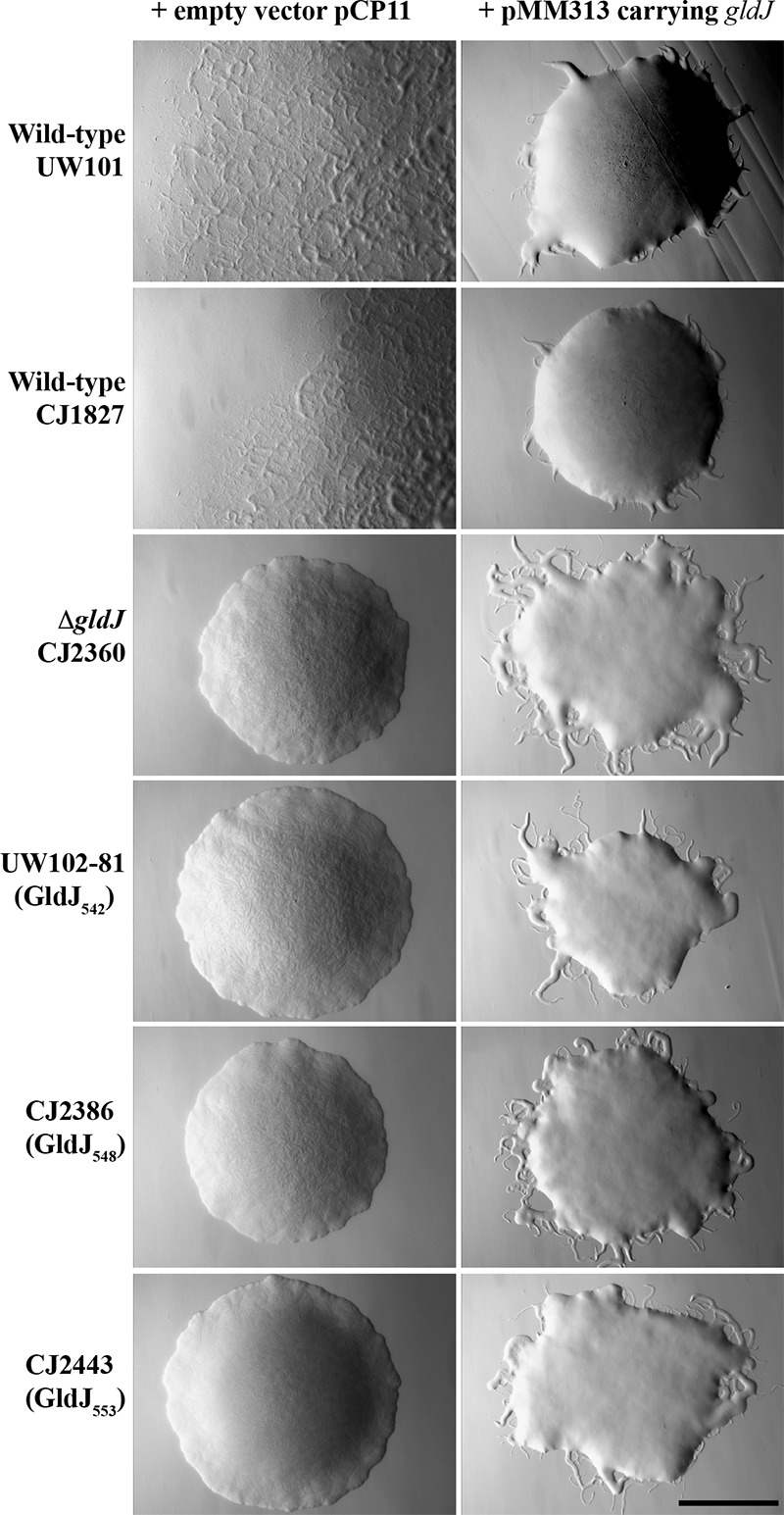
Photomicrographs of F. johnsoniae colonies. Colonies were incubated at 25°C on PY2 agar for 36 h. Photomicrographs were taken with a Photometrics Cool-SNAPcf2 camera mounted on an Olympus IMT-2 phase-contrast microscope. The wild-type strains used were F. johnsoniae UW101 and CJ1827 (rpsl mutant of UW101), and the ΔgldJ mutant was CJ2360. Truncated gldJ chromosomal mutants expressing GldJ proteins of 542 amino acids (GldJ542), 548 amino acids (GldJ548), and 553 amino acids (GldJ553) are also shown. The full-length wild-type GldJ is 561 amino acids in length. In each case, the colony on the left has the empty vector pCP11, and the colony on the right has pMM313, which expresses wild-type GldJ. The scale bar indicates 1 mm and applies to all panels.
FIG 13.
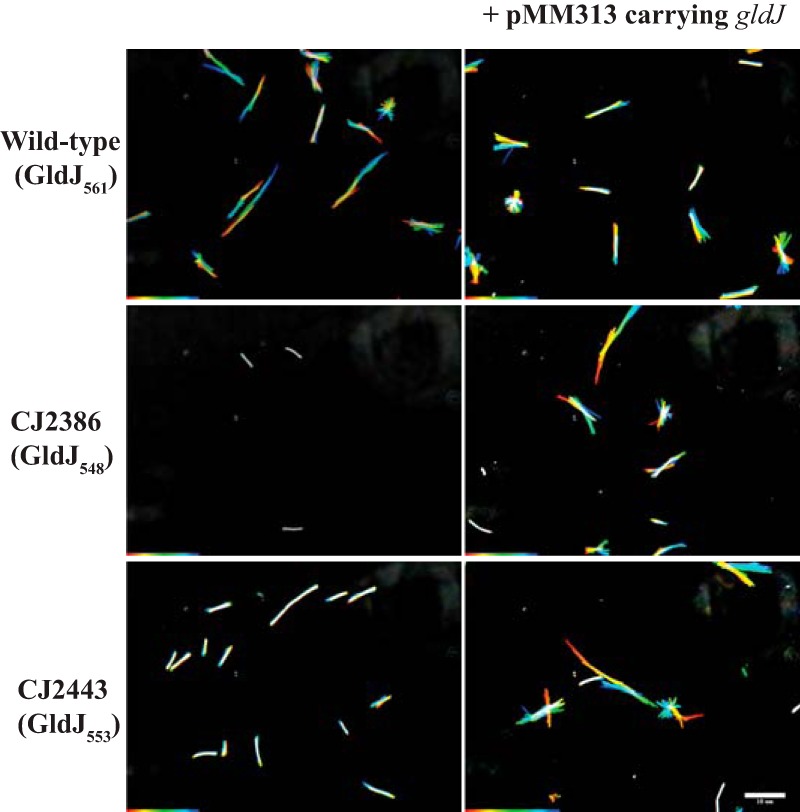
Gliding of wild-type cells and of cells of mutants that express truncated forms of GldJ. Cells were grown in MM overnight without shaking at 25°C, introduced into glass tunnel slides, and observed for motility at 25°C using a phase-contrast microscope. A series of images were taken for 30 s. Individual frames were colored from red (time zero) to yellow, green, cyan, and finally, blue (30 s), and integrated into one image, resulting in rainbow traces of gliding cells. White cells correspond to cells that exhibited little or no net movement. Multicolored “stars” indicate cells attached to the glass by one pole that rotated or flipped. The rainbow traces correspond to the sequences shown in Movies S2 and S3. Cells in the top row are wild-type F. johnsoniae CJ1827 and express full-length (561 aa) GldJ; the cells in the middle row are the gldJ mutant CJ2386, which expresses the first 548 aa of GldJ; and the cells in the bottom row are the gldJ mutant CJ2443, which expresses the first 553 aa of GldJ. Cells in the right column carry pMM313, which expresses full-length GldJ. The scale bar indicates 10 μm and applies to all panels.
The ability of strains expressing wild-type GldJ, GldJ548, and GldJ553 to propel SprB along the cell surface was examined. Wild-type cells expressing full-length GldJ561 bound and propelled anti-SprB-coated latex spheres, indicating the rapid movement of SprB (Movies S4 to S6). In contrast, cells expressing GldJ542 failed to bind the spheres (Fig. 10 and Movie S4), and cells expressing GldJ548 bound the spheres but failed to propel them (Fig. 10 and Movie S5). Cells expressing GldJ553 bound the anti-SprB-coated spheres and exhibited some slight movements of the spheres (Movie S6). Although many spheres failed to move, a few moved short distances, suggesting a severe but incomplete defect in motility. The introduction of pMM313 carrying wild-type gldJ restored the ability to propel SprB to each of the mutants (Movies S4 to S6). To explore the impact of the truncation of GldJ on SprB movement in more detail, spheres that were attached to different cells expressing full-length GldJ, GldJ548, and GldJ553 were examined quantitatively for movement (Table 1). Of the spheres attached to cells expressing full-length GldJ, 84% moved greater than 3 μm within 10 s, whereas 0% and 6% of the spheres attached to cells expressing GldJ548 and GldJ553 did. The few spheres that were propelled on cells expressing GldJ553 moved intermittently and more slowly than those on wild-type cells. As a result, when the window used to examine these cells was reduced to 5 s, none of the spheres attached to cells expressing GldJ553 moved 3 μm, whereas 76% of spheres on wild-type cells moved this distance. These results suggest that the C-terminal 13 amino acids of GldJ, while not required for the accumulation of GldK or for secretion of SprB by the T9SS, are important for the movement of SprB on the cell surface and for gliding motility.
TABLE 1.
Movement of latex spheres attached to SprB on wild-type and mutant cells
| Strain | GldJ protein expressed (aa) | % of spheres that moved >3 μm in 5- to 20-s observation perioda |
|||
|---|---|---|---|---|---|
| 5 s | 10 s | 15 s | 20 s | ||
| CJ1827 (wild type) | Full length (561) | 76 | 84 | 84 | 84 |
| CJ2386 | Truncated (548) | 0 | 0 | 0 | 0 |
| CJ2443 | Truncated (553) | 0 | 6 | 10 | 21 |
Movement of SprB was determined using anti-SprB antiserum and protein G-coated polystyrene spheres. The movement of 106, 89, and 103 spheres attached to different cells of strains CJ1827, CJ2386, and CJ2443, respectively, were examined. Spheres that moved along the cell surface a distance of greater than 3 μm were recorded at 5 s, 10 s, 15 s, and 20 s after the beginning of observation.
DISCUSSION
Many members of the phylum Bacteroidetes have gliding motility and T9SS machineries that are apparently not shared with bacteria outside this phylum (1). F. johnsoniae is a model organism for understanding gliding motility and T9SS-mediated protein secretion (9, 10, 15–17, 35). F. johnsoniae gliding motility and T9SS-mediated secretion both require the function of many novel proteins, and the two processes appear to be intertwined. More than 20 proteins involved in F. johnsoniae gliding motility have been identified (Fig. 1). Mutations in the genes encoding many of these proteins result in defects not only in gliding but also in chitin utilization and in susceptibility to bacteriophages (9, 10, 28, 32–34). Comparative analysis of F. johnsoniae with the nonmotile oral pathogen P. gingivalis provided an explanation for this pleiotropy and linked some of the gliding motility genes (gldK, gldL, gldM, gldN, sprA, sprE, and sprT) to protein secretion (9, 10, 15, 34). The proteins encoded by these genes are thought to form the core of the T9SS that secretes the motility adhesin SprB to the cell surface. This may explain the motility defects exhibited by T9SS mutants, since gliding involves the movement of SprB along the cell surface (12, 13). Circumstantial evidence suggested that mutations in some of the other motility genes (gldA, gldB, gldD, gldF, gldG, gldH, gldI, and gldJ) might also disrupt T9SS function. Mutations in these genes resulted not only in a loss of motility but also in an inability to digest chitin and resistance to bacteriophages (27–33). Since secretion of the extracellular chitinase ChiA requires the T9SS (16), and the T9SS is required for infection by bacteriophages (10), it was hypothesized that mutations in these 8 gld genes disrupted T9SS function. In this paper, we directly demonstrate that mutations in F. johnsoniae gldA, gldB, gldD, gldF, gldH, gldI, and gldJ result in T9SS defects, as shown by the inability of the mutants to secrete ChiA and SprB. We also present an explanation for the secretion defects, since the T9SS protein GldK was absent in each of the mutants. The mutants had normal levels of gldK mRNA, so a lack of transcription was not responsible for the lack of GldK protein. Instead, we suggest that the stability of the GldK protein may have been compromised in the mutants.
It was previously reported that strains with mutations in gldA, gldB, gldD, gldF, gldH, and gldI expressed gldJ mRNA but failed to accumulate the GldJ protein (28). This suggested that the proteins encoded by these genes were required for the stability of GldJ. Since GldJ is required for stable accumulation of the T9SS protein GldK, the other proteins (GldA, GldB, GldD, GldF, GldH, and GldI) may exert their effects on GldK levels through GldJ.
Truncated versions of GldJ lacking the C-terminal eight to 13 amino acids allowed an accumulation of GldK protein and thus, T9SS-mediated secretion of SprB, but failed to support efficient gliding motility. This observation defines a region of GldJ that is important for motility but not for secretion, and it begins to untangle these two processes. Comparative analysis of GldJ across 22 members of the phylum Bacteroidetes, most of which are known to exhibit gliding motility, revealed that regions near the C terminus are highly conserved (Fig. 8 and S2). The exact function of the C-terminal region in motility remains to be determined.
The gldJ truncation mutant expressing GldJ548 appeared to secrete SprB well but was partially defective for the secretion of ChiA (Fig. 9). There are several reasons that might explain this difference. The CTDs of ChiA and SprB that target them to the T9SS are not similar in sequence (16, 19). Further, ChiA requires PorV for its secretion, as do other proteins with type A CTDs (these typically belong to TIGRFAM family TIGR04183) (17, 19). In contrast, SprB has a type B CTD (belonging to family TIGR04131) and does not require PorV for its secretion; instead, it requires the unrelated protein, SprF (8, 17). Proteins with type A CTDs and proteins with type B CTDs both require the T9SS for secretion, but they appear to interact differently with this system. Further study is needed to determine if these differences result in the apparently more severe defect of secretion of ChiA by the gldJ truncation mutant producing GldJ548. At any rate, it is clear that SprB is secreted to the cell surface of truncated gldJ mutant cells expressing GldJ548 and GldJ553, and that these mutants are defective in gliding motility and in the movement of SprB along the cell surface. These results begin to separate T9SS function from gliding motility.
The results reported also reveal aspects of the role of the gld ABC transporter, composed of GldA, GldF, and GldG, in gliding. GldA, GldF, and GldG are required for accumulation of the GldJ protein (28), which, as shown here, is needed for accumulation of the T9SS component GldK. Not only is the GldA protein required for the accumulation of GldJ and GldK, but apparently, GldA function is also required. Mutations predicted to disrupt the ATP binding site of GldA resulted in a lack of accumulation of GldJ and GldK. The function of GldA in motility is not known, but it probably does not function as part of the motor, because the gliding of F. johnsoniae and related bacteria is dependent on proton motive force rather than on ATP hydrolysis (12, 36–39). Most gliding members of the phylum Bacteroidetes have orthologs of gldA, gldF, and gldG, and the majority of those that fail to glide (including P. gingivalis) lack orthologs (1). The cooccurrence of gldA, gldF, and gldG with gliding, although not universal, suggests that the encoded proteins may perform an important function in motility. However, at least two members of the Bacteroidetes that exhibit active gliding, Cellulophaga algicola and Maribacter sp. strain HTCC2170, lack orthologs of gldA, gldF, and gldG (1, 40). Thus, although these genes are required for F. johnsoniae gliding, they are not required for gliding of all members of the Bacteroidetes.
The motility machinery and T9SS of F. johnsoniae appear to be intertwined, which is reminiscent of the relationship between the bacterial flagellum and the type III secretion system associated with its assembly (41). We have begun to untangle Flavobacterium gliding motility from secretion, but the extent to which these processes can be separated remains unclear. The results presented here demonstrate that GldJ has a role in motility independent of its role in the secretion of SprB, but the exact function of GldJ in motility is not known. GldJ is an outer membrane lipoprotein and appears to localize in a helical manner (28). Given the helical movements of SprB along the cell surface and the helical movements of gliding cells (12, 42), it is possible that GldJ is a component of a structure on the periplasmic surface of the outer membrane on which SprB is propelled by the gliding motor. Further experiments are needed to clarify the exact role of GldJ in motility. Motility appears to be powered by the proton gradient across the cytoplasmic membrane (12, 38), and proteins that span this membrane must be involved in this process. Besides GldF and GldG (part of the ABC transporter described above), the only known motility proteins that span the cytoplasmic membrane are GldL and GldM. These are components of the T9SS and may drive protein secretion across the outer membrane. Recent results suggest that GldL and GldM may also function as the motor that propels SprB along the cell surface, resulting in cell movement (14). Alternatively, other proteins (yet to be identified) may perform this motor function. Further study is necessary to determine the exact nature of the Bacteroidetes gliding motor and its relationship to protein secretion.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
F. johnsoniae ATCC 17061T was the wild-type strain used in this study (43). Two versions of this wild-type strain (UW101 and FJ1) that both originated from the ATCC 17061T culture but that had slight phenotypic differences (32) were used in different experiments depending on the mutants under study, which were derived from strain UW101 or FJ1. The streptomycin-resistant rpsL mutant CJ1827 (derived from UW101) was used in some experiments, since this was the parent strain used to construct gene deletion mutants (44). F. johnsoniae cells were grown in Casitone-yeast extract (CYE) medium at 30°C, as previously described (45). To observe colony spreading, F. johnsoniae was grown at 25°C on PY2 medium supplemented with 10 g of agar per liter (27). Motility medium (MM) was used to observe the movement of individual cells in wet mounts (46). The strains and plasmids used in this study are listed in Table 2, and the primers are listed in Table S1 in the supplemental material. The plasmids used for complementation were all derived from pCP1 (45, 47) and have copy numbers of approximately 10 in F. johnsoniae. Antibiotics were used at the following concentrations when needed, unless indicated otherwise: ampicillin, 100 μg/ml; cefoxitin, 100 μg/ml; erythromycin, 100 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 20 μg/ml.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α MCR | Strain used for general cloning | Life Technologies (Grand Island, NY) |
| S17-1 λpir | Strain used for conjugation | 49 |
| HB101 | Strain used with pRK2013 for triparental conjugation | 50, 51 |
| F. johnsoniae | ||
| ATCC 17061T (UW101) | Wild type | 6, 32, 43 |
| ATCC 17061T (FJ1) | Wild type | 6, 32 |
| UW102-9 | gldA mutant encoding GldAG40R | 27 |
| UW102-41 | gldI-null mutant | 32 |
| UW102-55 | gldJ-null mutant | 28 |
| UW102-80 | gldJ-null mutant | 28 |
| UW102-81 | gldJ mutant encoding truncated GldJ of 542 amino acids | 28 |
| UW102-168 | gldA mutant encoding GldAR137C | 34 |
| CJ101-288 | gldA insertion mutant; (Emr) | 27 |
| CJ282 | gldD Tn4351 mutant; (Emr) | 31 |
| CJ569 | gldB Tn4351 mutant; (Emr) | 30 |
| CJ787 | gldF Tn4351 mutant; (Emr) | 29 |
| CJ1043 | gldH Tn4351 mutant; (Emr) | 33 |
| CJ1808 | chiA insertion mutant; (Emr) | 16 |
| CJ1827 | rpsL2; wild-type strain derived from ATCC 17061T (UW101) and used in construction of deletion mutants described below; (Smr) | 44 |
| CJ1922 | ΔsprB rpsL2 (Smr) | 44 |
| CJ2083 | remA::myc-tag-1 rpsL2 (Smr) | 11 |
| CJ2090 | ΔgldNO rpsL2 (Smr) | 11 |
| CJ2122 | ΔgldK rpsL2 (Smr) | 10 |
| CJ2157 | ΔgldL rpsL2 (Smr) | 10 |
| CJ2262 | ΔgldM rpsL2 (Smr) | 10 |
| CJ2302 | ΔsprA rpsL2 (Smr) | 10 |
| CJ2360 | ΔgldJ remA::myc-tag-1 rpsL2 (Smr) | This study |
| CJ2386 | gldJ548 rpsL2; gldJ deletion mutation that results in production of GldJ protein truncated after the 548th amino acid; (Smr) | This study |
| CJ2443 | gldJ553 rpsL2; gldJ deletion mutation that results in production of GldJ protein truncated after the 553rd amino acid; (Smr) | This study |
| FJ149 | sprE HimarEm2 mutant; (Emr) | 9 |
| KDF001 | sprT insertion mutant; (Emr) | 15 |
| Plasmids | ||
| pCP11 | E. coli-F. johnsoniae shuttle plasmid; Apr (Emr) | 45 |
| pCP23 | E. coli-F. johnsoniae shuttle plasmid; Apr (Tcr) | 27 |
| pCP29 | E. coli-F. johnsoniae shuttle plasmid; Apr (Cfr Emr) | 52 |
| pSA21 | pCP23 carrying gldA; Apr (Tcr) | 27 |
| pDH223 | pCP11 carrying gldB; Apr (Emr) | 30 |
| pMM209 | pCP23 carrying gldD; Apr (Tcr) | 31 |
| pMK314 | pCP29 carrying gldF and gldG; Apr (Cfr Emr) | 29 |
| pMM293 | pCP23 carrying gldH; Apr (Tcr) | 33 |
| pMM291 | pCP11 carrying gldI; Apr (Emr) | 32 |
| pMM313 | pCP11 carrying gldJ; Apr (Emr) | 28 |
| pTB99 | pCP23 carrying gldK; Apr (Tcr) | 6 |
| pRR51 | rpsl-containing suicide vector used for constructing deletion mutants; Apr (Emr) | 44 |
| pMM317 | Fragment encoding the first 337 amino acids of GldJ inserted in pCP11; Apr (Emr) | 28 |
| pMM318 | Fragment encoding the first 547 amino acids of GldJ inserted in pCP11; Apr (Emr) | 28 |
| pAB31 | Fragment encoding the first 542 amino acids of GldJ inserted in pCP23; Apr (Tcr) | This study |
| pJJ07 | Construct used to delete gldJ; 2.3-kbp upstream and 3.0-kbp downstream of gldJ amplified and ligated into pRR51; Apr (Emr) | This study |
| pJJ09 | 1.9-kbp region upstream of base 1644 of gldJ (numbered from the “A” of gldJ start codon) amplified with primers 1502 and 1503 and ligated into BamHI- and SalI-digested pRR51; Apr (Emr) | This study |
| pJJ10 | Construct used to generate truncated gldJ encoding the first 548 amino acids of GldJ (GldJ548); 1.9-kbp region downstream of gldJ amplified with primers 1504 and 1505 and ligated into SalI- and SphI-digested pJJ09; Apr (Emr) | This study |
| pJJ11 | Construct used to generate truncated gldJ encoding the first 553 amino acids of GldJ (GldJ553); 1.9-kbp region upstream of base 1659 of gldJ (numbered from the “A” of gldJ start codon) amplified with primers 1502 and 1555 and ligated into BamHI- and SalI-digested pJJ10; Apr (Emr) | This study |
Antibiotic resistance phenotypes are as follows: Apr, ampicillin resistance; Cfr, cefoxitin resistance; Emr, erythromycin resistance; Smr, streptomycin resistance; Tcr, tetracycline resistance. The antibiotic resistance phenotypes given in parentheses are those expressed in F. johnsoniae but not in E. coli. The antibiotic resistance phenotypes without parentheses are those expressed in E. coli but not in F. johnsoniae.
Determination of sites of gldA mutations.
The region spanning gldA was amplified from the F. johnsoniae gldA mutants UW102-9 and UW102-168 by PCR using primers 21 and 22. The PCR products were sequenced to identify the individual mutations.
Construction of the gldJ deletion mutant CJ2360.
An unmarked deletion of gldJ was constructed essentially as previously described (44). Briefly, a 2.3-kbp fragment upstream of gldJ was amplified by PCR using Phusion DNA polymerase (New England BioLabs, Ipswich, MA) and primers 1329 (introducing BamHI site) and 1330 (introducing a SalI site). The fragment was digested using BamHI and SalI and ligated into pRR51 that had been digested with the same enzymes, generating pJJ06. The 3.0-kbp fragment downstream of gldJ was amplified by PCR with primers 1331 (introducing SalI site) and 1332 (introducing SphI site). The fragment was digested with SalI and SphI and ligated into pJJ06 that had been digested with the same enzymes, generating the deletion construct pJJ07. Plasmid pJJ07 was introduced into F. johnsoniae strain CJ2083 by triparental conjugation, and deletion mutants were isolated as previously described (44).
Construction of strains expressing truncated forms of GldJ.
pMM317 and pMM318 produce truncated versions of GldJ that are 337 and 547 amino acids long, respectively (28). pAB31, which expresses the first 542 amino acids of GldJ, was generated by amplifying and cloning a fragment of gldJ into pCP23. Primers 1360 (introducing a BamHI site) and 1361 (introducing a stop codon and an SphI site) were used to amplify a region spanning the first 1,626 bp of gldJ. The product was digested with BamHI and SphI and inserted into pCP23 that had been digested with the same enzymes to generate pAB31. pMM317, pMM318, and pAB31 were transferred into the gldJ mutant UW102-55. UW102-55 has a frameshift mutation near the beginning of gldJ that allows only the first 29 amino acids of GldJ to be translated (28).
Strains with chromosomal mutations resulting in the production of truncated forms of GldJ were also identified and constructed. F. johnsoniae strain UW102-81, which was previously described, has a base substitution in gldJ (A to T at position 1627 numbered from the A of the gldJ start codon), resulting in the production of GldJ truncated after amino acid 542 (28, 48). Mutants expressing GldJ truncated after amino acids 548 and 553 were generated by markerless allele replacement, essentially as previously described (44). To delete the region encoding the C-terminal 13 amino acids, a 1.9-kbp fragment upstream of gldJ nucleotide 1644 (part of the R548 codon) was amplified by PCR using primers 1502 (introducing the BamHI site) and 1503 (introducing a stop codon, followed by the SalI site). The fragment was digested using BamHI and SalI and ligated into pRR51 that was digested with the same enzymes to generate pJJ09. The 1.9-kbp fragment downstream of gldJ was amplified by PCR with primers 1504 (introducing the SalI site) and 1505 (introducing the SphI site). The fragment was digested with SalI and SphI and ligated into pJJ09 digested with the same enzymes to generate the deletion construct pJJ10. Plasmid pJJ10 was introduced into streptomycin-resistant wild-type F. johnsoniae strain CJ1827 by triparental conjugation, and deletion mutants were isolated as previously described (44). Deletion of the C-terminal 8 amino acids was performed using pJJ11, which was constructed in the same way as pJJ10, except that primers 1502 and 1555 were used to amplify the 1.9-kbp fragment upstream of gldJ nucleotide 1659 (part of the T553 codon). All deletions were confirmed by DNA sequencing.
Measurement of chitin utilization.
The ability of F. johnsoniae to utilize chitin was assayed as described previously (34). F. johnsoniae cells were grown overnight in MM without shaking at 25°C. Two microliters of cells was spotted on MYA-chitin containing appropriate antibiotics, and plates were incubated for 2.5 days and examined for clearing zones.
Western blot analyses.
Wild-type and mutant cells of F. johnsoniae were grown to late-log phase in CYE at 25°C, except for analyses of ChiA, in which case cells were grown in MM at 25°C. Cells were washed twice in phosphate-buffered saline (PBS) consisting of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, and 2 mM KH2PO4 (pH 7.4) by centrifugation at 4,000 × g, and they were lysed by incubation for 5 min at 100°C in SDS-PAGE loading buffer. Proteins (15 μg per lane) were separated by SDS-PAGE, and Western blotting was performed as described previously (34), except that polyvinylidene difluoride (PVDF) membranes were used instead of nitrocellulose. Affinity-purified antisera (10, 13, 16, 28, 29, 34) were used to detect GldA, GldJ, GldK, GldL, GldM, GldN, SprB, and ChiA. Cell-associated and soluble secreted ChiA were separated as previously described (16). In brief, cells grown in MM were pelleted by centrifugation, and the culture supernatant (spent medium) was filtered using 0.22-μm-pore-size PVDF filters (Thermo Fisher Scientific, Rockford, IL). For whole-cell samples, the cells were suspended in the original culture volume of PBS. Equal amounts of spent medium and whole cells were boiled in SDS-PAGE loading buffer, proteins were separated by SDS-PAGE, and Western blot analyses were performed. Equal amounts of each sample based on the starting material were loaded in each lane. For cell extracts, this corresponded to 15 μg of protein, whereas for spent medium, this corresponded to the equivalent volume of spent medium that contained 15 μg of cell protein before the cells were removed.
qPCR to characterize gldK mRNA levels in gld mutants.
Total RNA was isolated from F. johnsoniae cells grown in CYE at 25°C to late-log phase by using an RNeasy minikit, as previously described (Qiagen) (10). The concentration and purity of RNA were assayed with an Eppendorf model 6131 BioPhotometer (Eppendorf, Hamburg, Germany) and normalized to 100 ng/μl by dilution with diethyl pyrocarbonate-treated water. Contaminant DNA was digested with RNase-free DNase. cDNA was generated using the SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA) using gene-specific primers. Real-time quantitative PCR (qPCR) was performed using the Applied Biosystems SYBR Select master mix (Thermo Fisher Scientific) in a DNA Engine Opticon 2 system (MJ Research, Waltham, MA), according to the manufacturer's instructions. Gene-specific primers for first-strand synthesis and for qPCR are listed in Table S1. Running conditions for qPCR were as follows: 50°C for 2 min, 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, and 64°C for 40 s. A SYBR green fluorescence reading (excitation, 470 to 505 nm; sensing, 523 to 543 nm) was taken following each cycle to monitor double-stranded DNA (dsDNA) product accumulation. A dissociation curve was generated to assay for single PCR product formation following the amplification phase by reading fluorescence every 0.2°C increase from 55°C to 98°C. All PCRs were carried out in triplicate with two independent qPCR experiments. The PCR products were also analyzed by agarose gel electrophoresis.
Microscopic observations of cell movement.
Wild-type and mutant cells were examined for movement over glass by phase-contrast microscopy. Cells were grown overnight in MM at 25°C without shaking, as previously described (46). Tunnel slides were constructed using double-stick tape, glass microscope slides, and glass coverslips, as previously described (11). Cells in growth medium were introduced into the tunnel slides, incubated for 5 min, and observed for motility using an Olympus BH2 phase-contrast microscope with a heated stage at 25°C. Images were recorded with a Photometrics CoolSNAPcf2 camera and analyzed using the MetaMorph software (Molecular Devices, Downingtown, PA).
Analysis of wild-type and mutant cells for surface localization and movement of SprB.
The secretion of SprB was examined using anti-SprB antiserum and protein G-coated polystyrene spheres, essentially as previously described (10). Briefly, cells were grown to late-exponential phase in MM at 25°C. Purified anti-SprB (1 μl of a 1:10 dilution of a 300-mg/liter stock), 0.5-μm-diameter protein G-coated polystyrene spheres (1 μl of a 0.1% stock preparation; Spherotech, Inc., Libertyville, IL), and bovine serum albumin (BSA) (1 μl of a 1% solution) were added to 7 μl of cells (approximately 5 × 108 cells per ml) in MM. The cells were introduced into a tunnel slide and examined by phase-contrast microscopy at 25°C. Samples (in triplicate) were observed 3 min after spotting, images were recorded for 30 s, and 100 randomly selected cells were examined for the presence of spheres that remained attached to the cells during this time to determine the percentage of cells that had surface-associated SprB. Sequences of images were also examined to determine the percentage of attached spheres that moved greater than 3 μm during 5 to 20 s of observation.
Surface localization of SprB was also examined by immunofluorescence microscopy. Cells were grown in MM at 25°C without shaking until late-exponential phase. One milliliter of cell culture was centrifuged at 1,000 × g for 8 min, and the cell pellet was suspended in 1 ml of PBS. The cells were pelleted by centrifugation, suspended in fresh PBS, centrifuged again, and suspended in 50 μl of PBS plus 2% BSA. Cells were exposed to 50 μl of polyclonal anti-SprB final bleed (13) for 30 min at room temperature, and the cells were pelleted (1,000 × g for 8 min), washed 3 times with 500 μl of PBS, suspended in 50 μl of PBS plus 2% BSA, and exposed to 5 μl of F(ab′) fragment of goat anti-rabbit IgG conjugated to Alexa Fluor 488 (2 mg/ml; Invitrogen, Carlsbad, CA). Cells were incubated for 30 min in the dark and collected by centrifugation. Cells were washed once, suspended in 500 μl of PBS, and kept in the dark until analysis (no more than 1 h after labeling). Samples were spotted on glass slides previously coated with a thin layer of 0.7% agarose and allowed to sit 30 s before applying coverslips. Samples were observed using a Nikon Eclipse 80i microscope with a LH-M100CB-1 mercury lamp and B-2E/C filter (Nikon Instruments, Inc., Melville, NY). Epifluorescence and differential interference contrast (DIC) images were recorded with a QImaging Retiga 2000R camera through a Nikon Plan Apo 40×/0.95 DIC M/N2 objective using the QCapture Pro software (QImaging, Surrey, British Columbia, Canada).
Fluorescent labeling of cells was quantified by flow cytometry. Cells were cultured as described above, collected by centrifugation, washed with PBS, and suspended in PBS at approximately 2 × 106 CFU/ml. Cells were labeled with anti-SprB and F(ab′) fragment of goat anti-rabbit IgG conjugated to Alexa Fluor 488, as described above. The fluorescence intensities were measured using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA), with a 488-nm laser. A minimum of 49,877 particles having the light-scattering properties of bacteria were analyzed for each sample, and fluorescence data were displayed using a 4-decade log scale. Growth media and buffers used for immunofluorescence microscopy and flow cytometry were filtered (0.22-μm pore size) before use.
Supplementary Material
ACKNOWLEDGMENTS
We thank Doug Steeber and Heather Owen for assistance with the flow cytometry and immunofluorescence microscopy experiments, respectively.
This research was supported by grants MCB-1021721 and MCB-1516990 from the National Science Foundation.
The funder had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00362-17.
REFERENCES
- 1.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J Bacteriol 195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarrell KF, McBride MJ. 2008. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 3.Islam ST, Mignot T. 2015. The mysterious nature of bacterial surface (gliding) motility: a focal adhesion-based mechanism in Myxococcus xanthus. Semin Cell Dev Biol 46:143–154. doi: 10.1016/j.semcdb.2015.10.033. [DOI] [PubMed] [Google Scholar]
- 4.Miyata M. 2010. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol 64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 5.Nan B, Zusman DR. 2016. Novel mechanisms power bacterial gliding motility. Mol Microbiol 101:186–193. doi: 10.1111/mmi.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol 187:6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson SS, Glocka PP, Agarwal S, Grimm DP, McBride MJ. 2007. Flavobacterium johnsoniae SprA is a cell-surface protein involved in gliding motility. J Bacteriol 189:7145–7150. doi: 10.1128/JB.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhodes RG, Nelson SS, Pochiraju S, McBride MJ. 2011. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J Bacteriol 193:599–610. doi: 10.1128/JB.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhodes RG, Samarasam MN, Van Groll EJ, McBride MJ. 2011. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J Bacteriol 193:5322–5327. doi: 10.1128/JB.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. 2013. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell-surface gliding motility adhesins SprB and RemA. J Bacteriol 195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. 2012. Flavobacterium johnsoniae RemA is a mobile cell-surface lectin involved in gliding. J Bacteriol 194:3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. 2013. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A 110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson SS, Bollampalli S, McBride MJ. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol 190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrivastava A, Lele PP, Berg HC. 2015. A rotary motor drives Flavobacterium gliding. Curr Biol 25:338–341. doi: 10.1016/j.cub.2014.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kharade SS, McBride MJ. 2014. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol 196:961–970. doi: 10.1128/JB.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharade SS, McBride MJ. 2015. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol 197:147–158. doi: 10.1128/JB.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veith PD, Nor Muhammad NA, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification and cell surface attachment. J Proteome Res 12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 19.Kulkarni SS, Zhu Y, Brendel CJ, McBride MJ. 2017. Diverse C-terminal sequences involved in Flavobacterium johnsoniae protein secretion. J Bacteriol 199:e00884-. doi: 10.1128/JB.00884-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen KA, Travis J, Potempa J. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J Bacteriol 189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 22.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, Chen YY, Glew MD, Dashper SG, Reynolds EC. 2015. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog 11:e1005152. doi: 10.1371/journal.ppat.1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YY, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O'Brien-Simpson N, Dashper SG, Reynolds EC. 2011. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol Microbiol 79:1380–1401. doi: 10.1111/j.1365-2958.2010.07530.x. [DOI] [PubMed] [Google Scholar]
- 25.Gorasia DG, Veith PD, Hanssen EG, Glew MD, Sato K, Yukitake H, Nakayama K, Reynolds EC. 2016. Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathog 12:e1005820. doi: 10.1371/journal.ppat.1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent MS, Canestrari MJ, Leone P, Stathopulos J, Ize B, Zoued A, Cambillau C, Kellenberger C, Roussel A, Cascales E. 2017. Characterization of the Porphyromonas gingivalis type IX secretion trans-envelope PorKLMNP core complex. J Biol Chem 292:3252–3261. doi: 10.1074/jbc.M116.765081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agarwal S, Hunnicutt DW, McBride MJ. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci U S A 94:12139–12144. doi: 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun TF, McBride MJ. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J Bacteriol 187:2628–2637. doi: 10.1128/JB.187.8.2628-2637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunnicutt DW, Kempf MJ, McBride MJ. 2002. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J Bacteriol 184:2370–2378. doi: 10.1128/JB.184.9.2370-2378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunnicutt DW, McBride MJ. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes, gldB and gldC. J Bacteriol 182:911–918. doi: 10.1128/JB.182.4.911-918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunnicutt DW, McBride MJ. 2001. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes gldD and gldE. J Bacteriol 183:4167–4175. doi: 10.1128/JB.183.14.4167-4175.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride MJ, Braun TF. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J Bacteriol 186:2295–2302. doi: 10.1128/JB.186.8.2295-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride MJ, Braun TF, Brust JL. 2003. Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J Bacteriol 185:6648–6657. doi: 10.1128/JB.185.22.6648-6657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhodes RG, Samarasam MN, Shrivastava A, van Baaren JM, Pochiraju S, Bollampalli S, McBride MJ. 2010. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J Bacteriol 192:1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride MJ. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu Rev Microbiol 55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 36.Duxbury T, Humphrey BA, Marshall KC. 1980. Continuous observations of bacterial gliding motility in a dialysis microchamber: the effects of inhibitors. Arch Microbiol 124:169–175. doi: 10.1007/BF00427723. [DOI] [Google Scholar]
- 37.Dzink-Fox JL, Leadbetter ER, Godchaux W III. 1997. Acetate acts as a protonophore and differentially affects bead movement and cell migration of the gliding bacterium Cytophaga johnsonae (Flavobacterium johnsoniae). Microbiology 143:3693–3701. doi: 10.1099/00221287-143-12-3693. [DOI] [PubMed] [Google Scholar]
- 38.Pate JL, Chang L-YE. 1979. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr Microbiol 2:59–64. doi: 10.1007/BF02601737. [DOI] [Google Scholar]
- 39.Ridgway HF. 1977. Source of energy for gliding motility in Flexibacter polymorphus: effects of metabolic and respiratory inhibitors on gliding movement. J Bacteriol 131:544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y, McBride MJ. 2016. Comparative analysis of Cellulophaga algicola and Flavobacterium johnsoniae gliding motility. J Bacteriol 198:1743–1754. doi: 10.1128/JB.01020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macnab RM. 2004. Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta 1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Shrivastava A, Roland T, Berg HC. 2016. The screw-like movement of a gliding bacterium is powered by spiral motion of cell-surface adhesins. Biophys J 111:1008–1013. doi: 10.1016/j.bpj.2016.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, Staroscik AM, Hoover TR, Cheng YQ, Stein JL. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol 75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodes RG, Pucker HG, McBride MJ. 2011. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant motility genes remF, remG, remH, and remI. J Bacteriol 193:2418–2428. doi: 10.1128/JB.00117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride MJ, Kempf MJ. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae J Bacteriol 178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, McBride MJ, Subramaniam S. 2007. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol 189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alvarez B, Secades P, McBride MJ, Guijarro JA. 2004. Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum. Appl Environ Microbiol 70:581–587. doi: 10.1128/AEM.70.1.581-587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang LYE, Pate JL, Betzig RJ. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J Bacteriol 159:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Lorenzo V, Timmis KN. 1994. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 50.Bolivar F, Backman K. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol 68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- 51.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kempf MJ, McBride MJ. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J Bacteriol 182:1671–1679. doi: 10.1128/JB.182.6.1671-1679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



