Fig. 2.
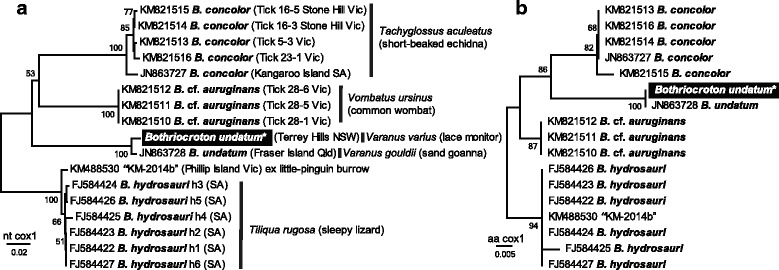
Phylogenetic position of the Goanna tick (Bothriocroton undatum). Ticks were collected from a wild lace monitor (Varanus varius) from Sydney, Australia. a The evolutionary history based on cox1 nucleotide (nt) sequences was inferred using the Minimum Evolution (ME) method with Kimura-2 parameter model and percent bootstrap support percentages (200 replicates) shown next to the branches was calculated in MEGA7. There were a total of 521 positions in the final dataset. b The evolutionary history based on cox1 amino acid (aa) sequences was inferred using the ME method, with bootstrap support percentages test (200 replicates) shown next to the branches. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of aa substitutions per site. There were a total of 173 positions in the final dataset. Accession number with tick species name, tick identifier and locality in Australia is followed by host name. *All B. undatum ticks (n = 6, Tick1 to Tick6) had identical cox1 sequences
