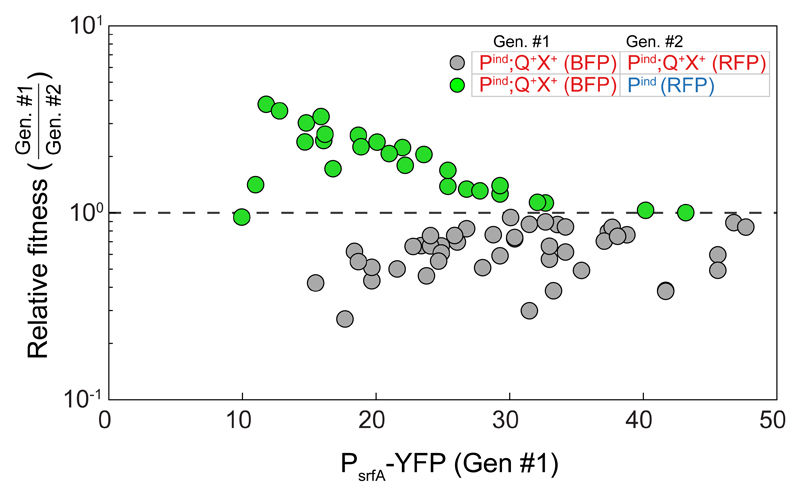
Figure 3. Self-sensing contributes to antibiotic persistence.
The relative fitness of co-cultured strains is plotted as a function of the PsrfA-YFP expression of genotype #1 prior to administration of antibiotics (see legend). Relative fitness was calculated as the ratio of relative frequency of the strains at the end of the experiment to that prior to the addition of antibiotics [19]. Shown are results for two differentially marked Pind;Q+X+-secreting strains (gray), and between the physiological ComX-secreting (Pind;Q+X+) and the corresponding non-secreting (Pind) strains (green). A value of one (dashed line) indicates no change in frequency. Each data point represents a separate measurement. Data was collected over ≥3 different optical densities in each series of experiments. Experiments were repeated multiple times over ≥3 days for each co-culture type. All data points are given in Supplementary File 1.