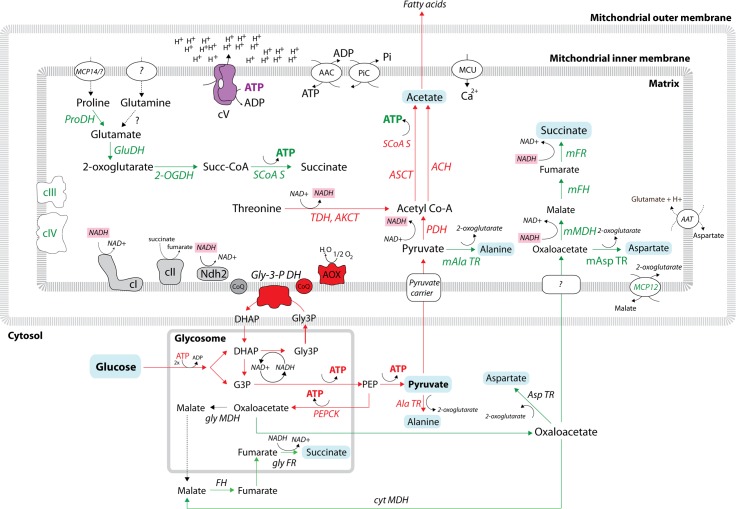
Fig 2. Schematic representation of carbon source metabolism in the bloodstream form of T. brucei.
Red arrows represent enzymatic steps that were experimentally shown to be active in BSF. Green arrows represent enzymatic steps that might be active in BSF because the enzymes (in green) were identified in BSF proteomic data. Glucose-derived metabolites (acetate, pyruvate, succinate, alanine, aspartate) are on a blue background. NADH molecules are on a pink background. Dashed arrows indicate enzymatic steps for which no experimental proof exists. The glycosomal and mitochondrial compartments are indicated. 2-OGDH, 2-oxoglutarate dehydrogenase; AAC, ADP/ATP carrier; AAT, amino acid transporter; ACH, acetyl-CoA thioesterase; AKCT, 2-amino-3-ketobutyrate coenzyme A ligase; Ala TR, alanine transaminase; AOX, alternative oxidase; ASCT, acetate:succinate CoA-transferase; Asp TR, aspartate transaminase; BSF, bloodstream form; cI, complex I (NADH:ubiquinone oxidoreductase); cII, complex II (succinate dehydrogenase); cIII, complex III (cytochrome bc1 complex); cIV, complex IV (cytochrome c oxidase); cV, complex V (FoF1 ATPase); cyt, cytosolic; DHAP, dihydroxyacetone phosphate; FH, fumarate hydratase (i.e., fumarase); FR, fumarate reductase; G3P, glyceraldehyde 3-phosphate; GluDH, glutamate dehydrogenase; gly, glycosomal; Gly3P, glycerol 3-phosphate; Gly-3-PDH, glycerol-3-phosphate dehydrogenase; m, mitochondrial; MDH, malate dehydrogenase; PDH, pyruvate dehydrogenase; PEP, phosphoenolpyruvate; PEPCK, phosphoenolpyruvate carboxykinase; PiC, phosphate carrier; ProDH, proline dehydrogenase.