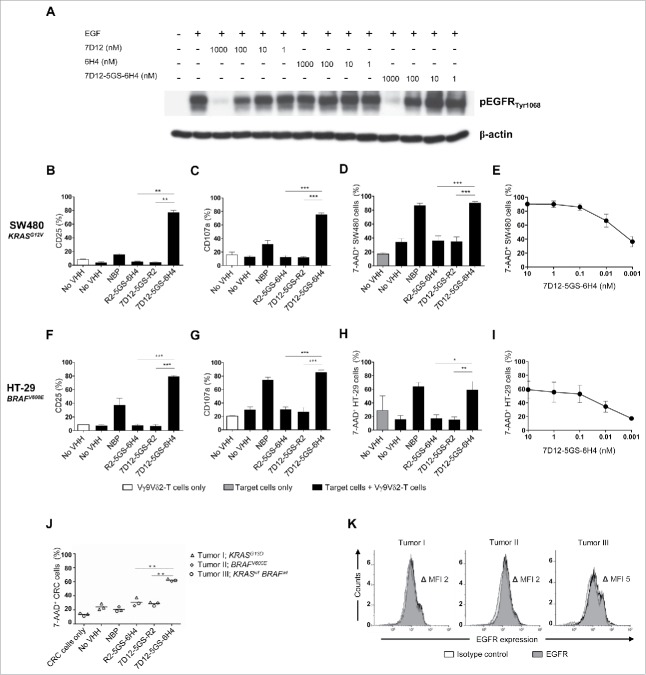
Figure 4.
The 7D12-5GS-6H4 bispecific VHH inhibits EGFR signaling but does not depend on this to induce tumor cell lysis. (A) The anti-EGFR 7D12 VHH retains its capacity to inhibit phosphorylation of EGFR when incorporated in a bispecific 7D12-5GS-6H4 VHH format. Her14 cells were incubated with a mixture of 8 nM human EGF and the indicated VHH. Cell lysates were run on SDS-PAGE gel and western blotted for phosphorylated EGFRTyr1068 and β-actin as a loading control. (B-I) Vγ9Vδ2-T cells were cultured with or without the EGFR+ colon tumor cells SW480 KRASG12V (B-E) or HT29 BRAFV600E (F-I) for 24 hours or primary colon cancer cells for 4 hours (J) in a 1:1 ratio in the presence of the 7D12-5GS-6H4 bispecific VHH or a bispecific control VHH. VHH concentrations: B-D, F-H and J) 10 nM; E and I) as indicated. For control conditions, Vγ9Vδ2-T cells were co-cultured with target cells in the absence of VHH (no VHH; negative control) or with NBP-pretreated target cells (positive control). Vγ9Vδ2-T cell activation and degranulation was determined by assessing the percentage of CD25 (B) and (F) or CD107a (C and G) expression, respectively by flow cytometry. The percentage of lysed target cells was determined using 7-AAD staining and flow cytometry (D-E and H-J). B-D and (F-H) White bars represent Vγ9Vδ2-T cell mono-cultures in the absence of VHH, grey bars represent target cell mono-cultures in the absence of VHH and black bars represent Vγ9Vδ2-T cell co-cultures with target cells and indicated VHH. E and (I) Co-cultures of target cells with Vγ9Vδ2-T cells and the indicated amount of 7D12-5GS-6H4 bispecific VHH. J) Lysis of patient derived primary colon cancer cells; tumor 1: mutation in KRAS exon 2, c.38G>A, p.G13D; tumor 2: mutation in BRAF exon 15, c.1799 T>A, p.V600E; tumor 3: RAS and BRAF wild-type. (K) All three colon cancer samples expressed EGFR as determined by flow cytometry. Shown are means ± SEM of n = 3 experiments. p-Values were calculated with a one-way ANOVA and Bonferroni's post-hoc test (* indicates p<0.05, ** indicates p<0.01 and *** indicates p<0.001). Abbreviations: aminobisphosphonate (NBP); Gly4Ser (GS); mean fluorescence index (MFI).