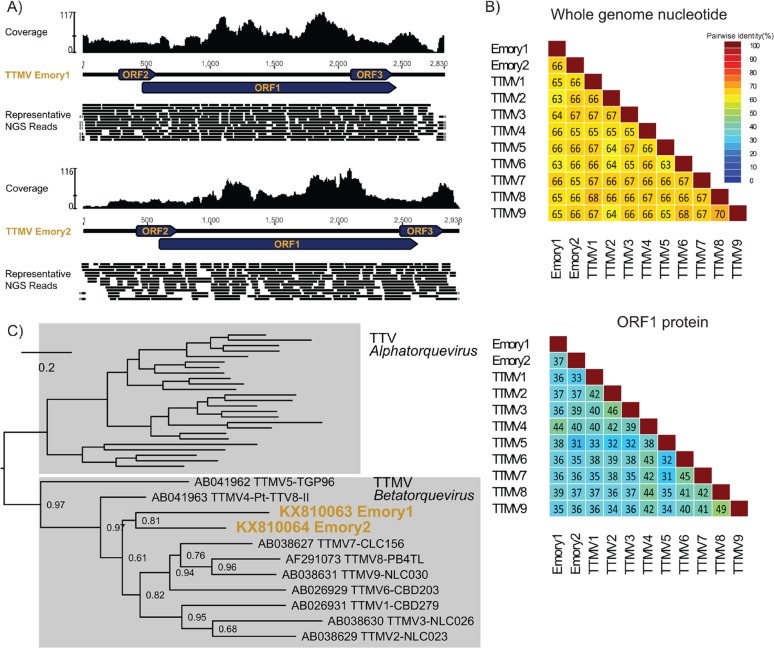
Figure 2. Comprehensive genomic and phylogenetic analysis of the Torque teno mini virus Emory1 (TTMV Emory1) and Torque teno mini virus Emory2 (TTMV Emory2).
(A) Schematic depiction of the TTMV Emory1 and Emory2 genome organization. Coverage for NGS data is indicated, and a portion of the representative NGS reads is shown. The circular genome is graphically linearized for display purpose, and the reads covering across the ends are marked with grey arrows on the extremities. (B) Pairwise whole genome nucleotide identities and ORF1 amino acid identities between Torque teno mini viruses. Protein sequences were aligned with MUSLE [52], and sequence identities were calculated using the species demarcation tool [53]. (C) ORF1 protein phylogeny of the genus Betatorquevirus, including the newly described TTMV Emory1 and Emory2. Maximum likelihood phylogeny was generated with PhyML [54], where branch support was calculated using Approximate Likelihood-Ratio Test (aLRT). The scale bar represents evolutionary distance in substitutions per site.