Fig. 4.
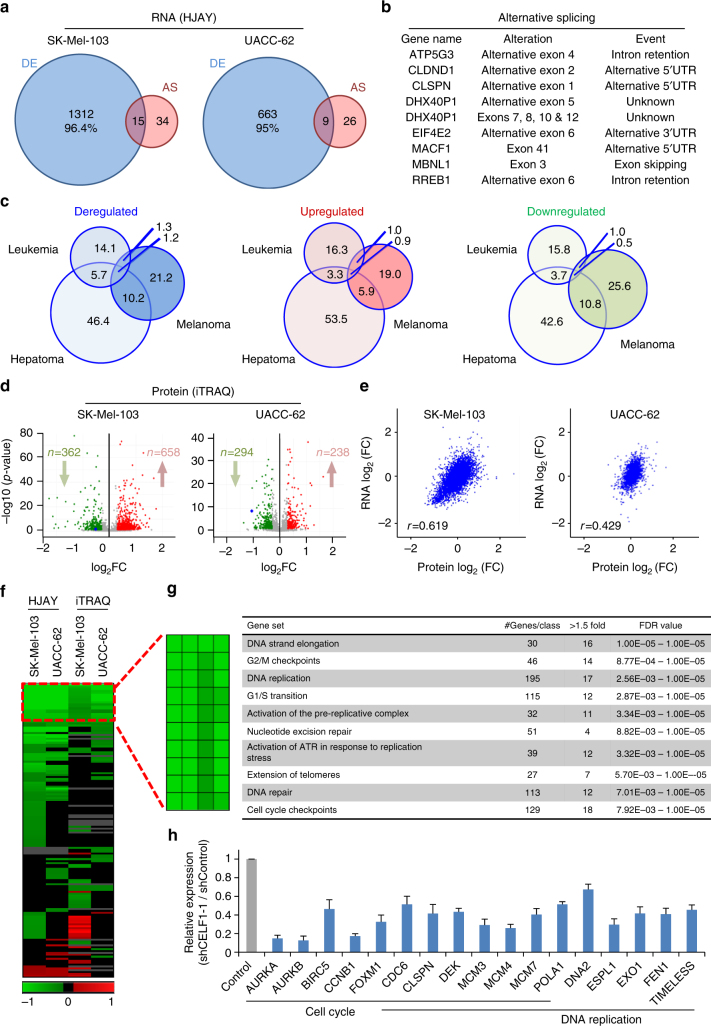
Genome-wide HJAY and iTRAQ LC–MS/MS identify new CELF1-regulated targets. a Differentially expressed (DE) and alternatively spliced (AS) genes identified by HJAY in SK-Mel-103 or UACC-62 cells transduced with shCELF1-1 estimated with respect to shRNA controls. The specific overlap in the mRNA expression changes is shown in Supplementary Fig. 7c, d. b Splicing alterations identified by the HJAY in SK-Mel-103 and UACC-62 upon CELF1 depletion. c Overlap (%) in changes in mRNA expression after depletion of CELF1 in melanoma cells (SK-Mel-103 and UACC-62) with respect to similar transcriptomic data available for K562 leukemia and HepG2 hepatoma cell lines (ENCODE ENCSR605MFS and ENCSR695XOD, respectively). The diameter of the Venn diagrams is proportional to the genes with mRNA changes in each data set. d Up- and downregulated proteins identified by iTRAQ LC–MS/MS analyses in SK-Mel-103 or UACC-62 cells upon shCELF1-1 transduction estimated with respect to shRNA controls. Up- and downregulated proteins (red and green, respectively) are shown as volcano plots. Factors indicated in green and red are those with significant changes in protein expression. Non significant changes in expression are labeled in gray (additional information for the relative overlap in the iTRAQ data for the cell lines analyzed is summarized in Supplementary Fig. 7e, f). e Correlation of global changes in RNA and protein levels upon CELF1 depletion in SK-Mel-103 and UACC-62 melanoma cell lines. f Heatmap of gene sets found enriched (Reactome pathway database) both at RNA and protein levels in shCELF1-1 expressing melanoma cells, listed as a function of FDR values. g Top-10 downregulated cellular functions identified in the two cell lines analyzed. Listed are enriched gene sets, the total number of genes in each category, the number of genes deregulated in both cell lines, and the range of FDR values (<0.25). h Expression levels of representative modulators of cell cycle and/or DNA replication upon CELF1 depletion in UACC-62 cells, as validation of data obtained from HJAY. Error bars correspond to SEM of three experiments in triplicate