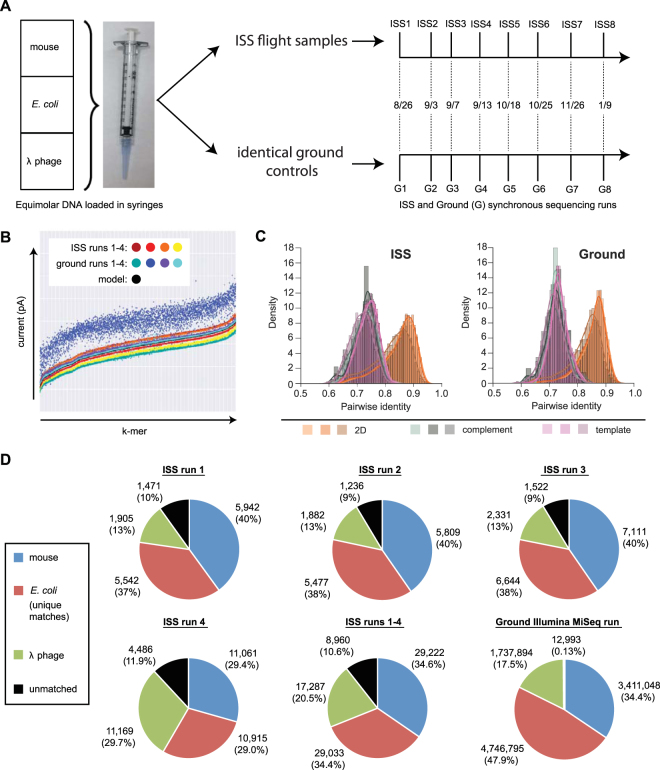
Figure 1.
Study design and flight/ground nanopore performance. (A) A mixture of equimolar DNA from mouse, E. coli and lambda phage genomes was sequenced in parallel on Earth (“Ground”) and in-flight on the ISS (after being delivered by a SpaceX Dragon capsule; flow cells were shipped to the Kennedy Space Center on July 11, 2016). Synchronous nanopore sequencing runs were performed from August 26, 2016 to January 9, 2017. (B) Plot of mean current intensity in picoAmperes (pA; Y-axis) against k-mers (x-axis) in order of increasing mean current based on a model distribution from Oxford Nanopore Technologies (black). Current distributions are tightly clustered with the exception of lower-quality ground #2. (C) Comparison of pairwise identities of aligned reads between ISS runs 1–4 and ground runs 1–4. (D) Pie charts of the read distributions corresponding to each ISS run and pooled ISS runs 1–4.