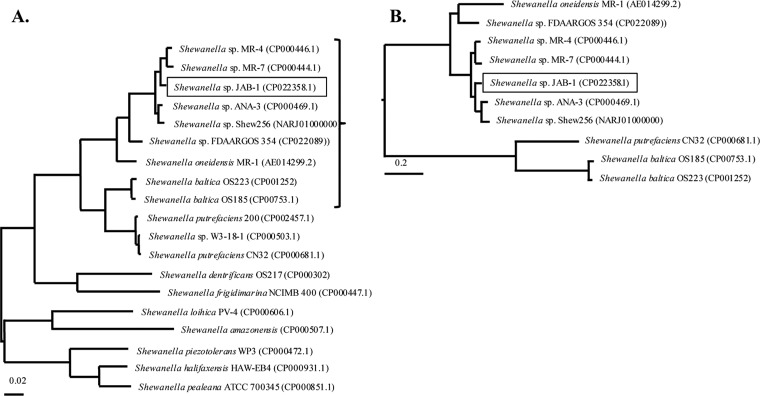
FIG 1.
Phylogenetic analysis of representative Shewanella spp. (A) Unrooted phylogenetic tree based on gyrB and rpoB genes. The tree was constructed by maximum likelihood method with Tamura-Nei model using MEGA program (MEGA7.0). The tree is drawn to scale, with branch lengths representing the evolutionary distances. (B) A focus on Shewanella spp. close to Shewanella sp. JAB-1 was made, and the phylogenetic tree was based on whole-genome sequences. It was constructed by maximum likelihood method with Jukes-Cantor model, using the Parsnp software. The tree is drawn to scale based on the single nucleotide polymorphisms (SNPs) of the alignment. Accession numbers are shown in parentheses next to each organism name.