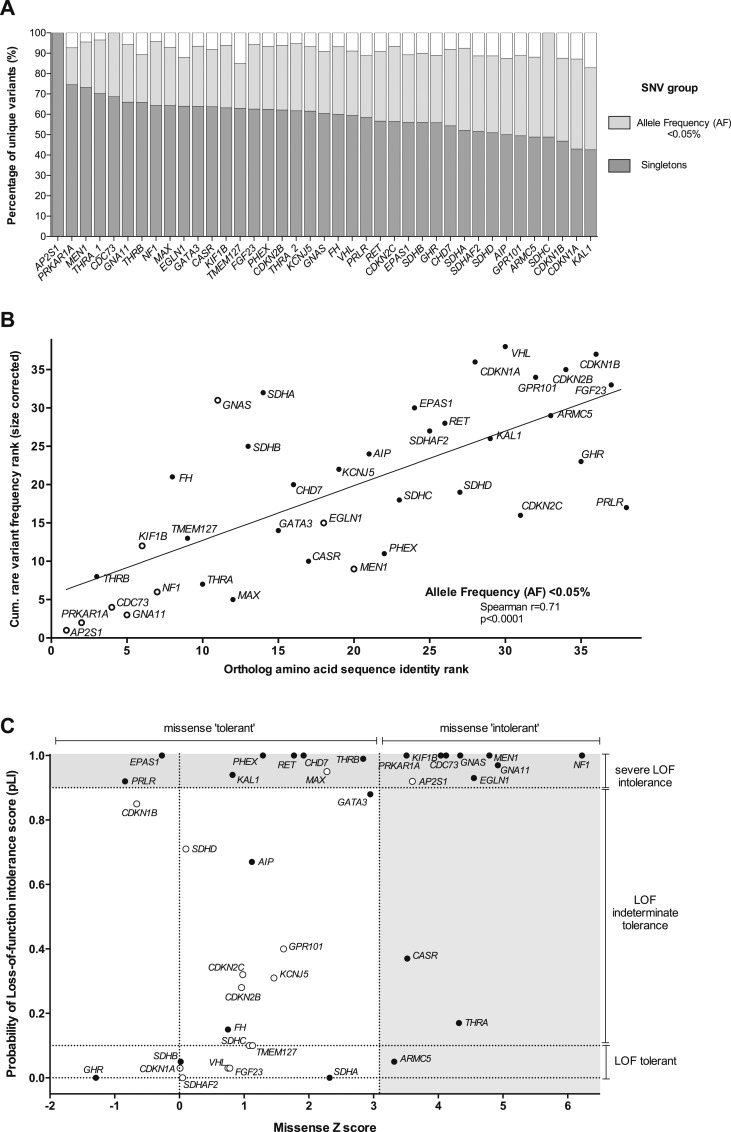
Figure 1.
Rare variant frequency, evolutionary conservation, and constraint metrics of genes associated with hereditary endocrine disease. (A) Proportion of all nonsynonymous SNVs occurring in the selected genes as singletons or with an AF <0.05%. Across the 38 genes, an average of 59.8% (range, 42.6% to 100%) of individual missense/LOF SNVs occurred as singletons, whereas 91.8% (range, 83% to 100%) had an AF <0.05%. (B) Rare SNV frequency was correlated with evolutionary conservation of the encoded protein. Individual genes were ranked according to both their size-corrected cumulative SNV frequency (for SNVs with AF <0.05%) and the degree of amino acid conservation between human and zebrafish (Danio rerio) orthologs. A significant correlation was observed (r = 0.69; P < 0.0001), such that genes with a high degree of conservation harbored the lowest rates of rare SNVs. Of note, a marked overlap was observed between genes with high conservation/low variation and those categorized as intolerant of both missense/LOF variation using constraint metrics (genes marked with open circles). All other genes are represented by closed circles. (C) Missense and LOF constraint metrics for the study genes. A z score >3.09 is reported to represent significant missense intolerance, whereas a pLI score >0.9 is indicative of extreme LOF intolerance and suggestive of a haploinsufficient function (i.e., gene intolerant to heterozygous LOF). Of note, 45% (17 of 38) of study genes could be classified as extreme LOF intolerant, whereas 32% (12 of 38) were missense intolerant. Several genes in which both missense and LOF mutations were responsible for penetrant monogenic disorders (e.g., MEN1, CDC73, and NF1) clustered in the combined LOF/missense intolerant group. In contrast, several genes in which the role of heterozygous germline variation in disease pathogenesis was less well defined were categorized as LOF (pLI score <0.1) and/or missense tolerant (e.g., CDKN1A, SDHA, and GPR101). However, the reliability of the pLI constraint metric was reduced for genes of small size where few LOF variants were predicted (e.g., <10), and these genes are identified by an open circle (e.g., CDKN1B, VHL). All other genes are represented by closed circles. pLI, probability of LOF intolerance.