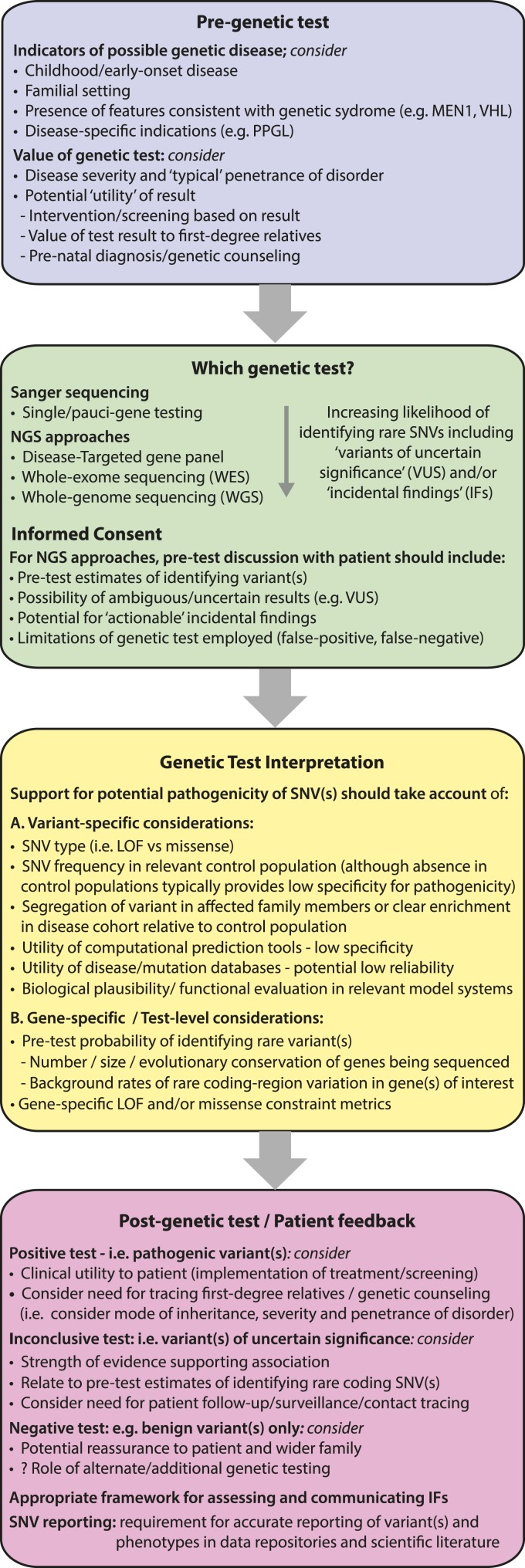
Figure 3.
Illustrative workflow outlining considerations for genetic testing and variant interpretation in clinical and research settings. Evaluating the spectrum and frequency of rare variations in large population cohorts may enhance the process of genetic testing at different stages of the clinical workflow. For example, as the content of genetic testing increases, the likelihood of identifying rare coding variants increases (e.g., VUSs and IFs). The current study illustrates how gene-specific cumulative rare variant frequencies may be used to establish pretest estimates for identifying such variants. This information could be incorporated into the informed consent process to alert patients to the likelihood of ambiguous test results. Although guidelines exist to standardize the process of ascribing pathogenicity to germline variants, these typically focus on variant-specific features. The current study highlights how gene-level factors, including estimates of rare missense/LOF variation together with metrics of constraint, may aid variant classification. For example, when the cumulative frequency of LOF variants in the control population exceeds the prevalence of the disease under investigation, the likelihood that such a variant is a high penetrance disease-allele is substantially reduced. Conversely, variants in genes with very low rare variation burden potentially have a higher likelihood of pathogenicity. In the future, it is possible that this information may contribute to Bayesian models for disease in which the likelihood of variant pathogenicity and/or disease expression is adjusted according to clinical factors as well as gene- and variant-level metrics. Furthermore, for potential disease alleles, refined estimates of disease penetrance may be established by evaluating the frequency of the variant in disease and control populations, and such accurate estimates are essential for appropriate genetic counseling (e.g., to determine the value of implementing treatment/surveillance programs and/or screening of first-degree relatives). In addition, when ambiguous test results have been obtained (e.g., VUSs), further refinement of risk may be established by relating the test result to both the pretest estimate of detecting such variation and constraint metrics, which together may aid the clinician and patient in making informed decisions regarding future care. Finally, these studies illustrate the need for transparent and accurate reporting of genetic data coupled to phenotypes (i.e., avoiding positive reporting bias) to improve the accuracy of existing disease/mutation databases. IF, incidental finding; NGS, next-generation sequencing; VUS, variant of uncertain significance.