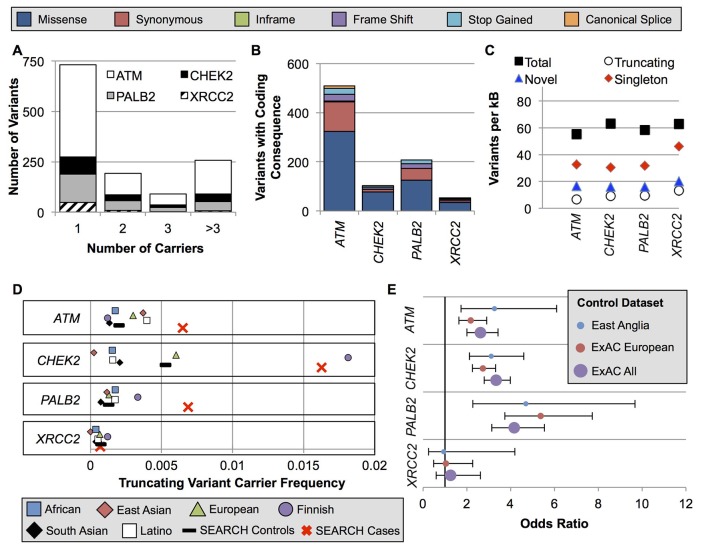
Figure 1.
Spectrum of ATM, CHEK2, PALB2 and XRCC2 variation in ExAC populations and East Anglian cases and controls. (A) Nearly all variants were very rare or private. (B) The absolute number of variants per gene was highly divergent due to the difference in gene size. (C) However, when corrected for gene size, the genes had similar per-kB rates of total, truncating, singleton and novel variants not previously reported in the dbSNP, ExAC or COSMIC. (D) The carrier frequency for truncating variants in the study controls was near the mean of the ExAC populations. Notably, the ExAC Finnish population was an outlier for CHEK2 and PALB2 due to well-studied founder effects. (E) OR point estimates were similar using the study controls versus ExAC populations as controls. ExAC, Exome Aggregation Consortium.