Figure 2.
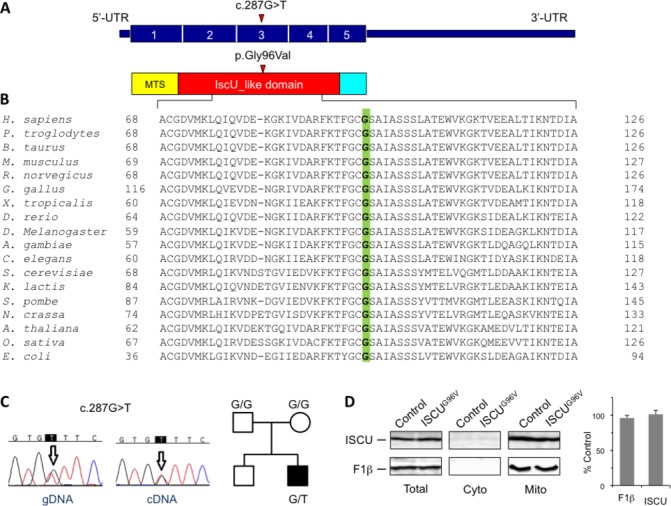
Identification and characterisation of an ISCU mutation. (A) Schematic representation of the ISCU cDNA (NM_213595.2) and ISCU protein with the nucleotide/amino acid change identified in this study. The functional IscU-like domain is in red; the mitochondrial targeting sequence (MTS) is in yellow. (B) Phylogenetic conservation of the amino acid residue (Gly96, in green) affected by the missense mutation identified in the patient. (C) Electropherograms of the genomic region (gDNA) and transcript (cDNA) harbouring the ISCU mutation, and pedigree. (D) Immunoblot analysis of ISCUG96V mutant protein expression and subcellular localisation. Control and patient fibroblasts were harvested by trypsination, permeabilised by digitonin treatment, and separated into a cytosolic and a mitochondria-containing membrane fraction. Total cell lysates as well as cytosolic and crude mitochondrial fractions were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analysed for ISCU and ATP synthase F1β subunit (mitochondrial marker) steady-state protein levels (left panel). Chemiluminescence signals of ISCU and F1β in total lysate samples were quantified, and values obtained from patient fibroblasts were expressed relative to control cells (right panel). Error bars indicate the SDs (n=3). UTR, untranslated region.
