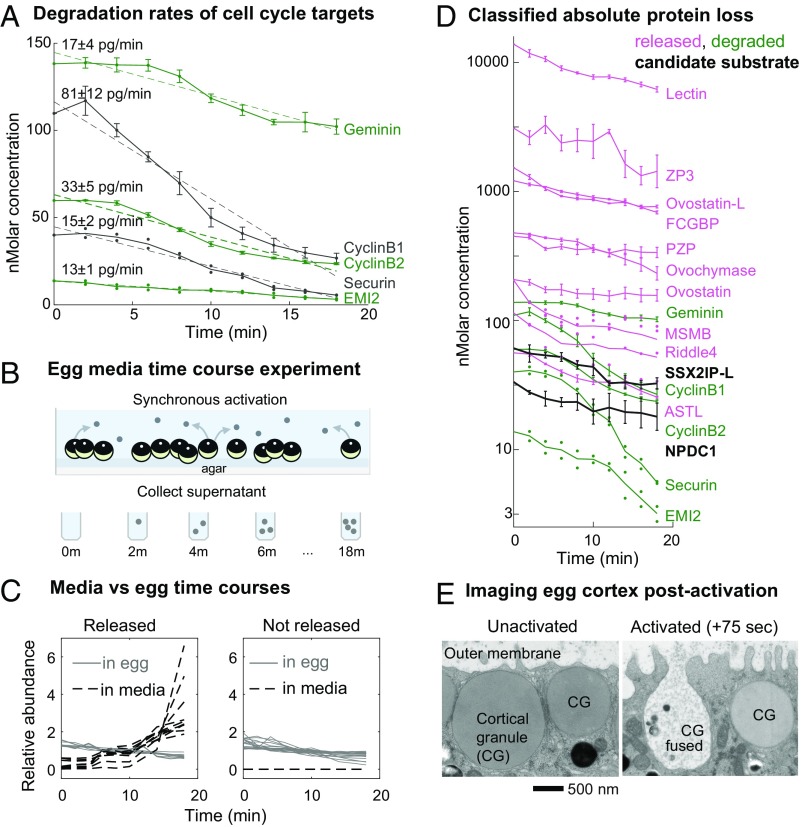
Fig. 2.
Protein loss occurs by degradation and release: Experiments and analysis performed to classify the mechanism of protein loss following egg activation. (A) Time series of the five proteins identified as significantly decreasing, which are known cell cycle degradation targets, plotted by their absolute changes. Error bars reflect SE (SEM) for proteins detected in at least three biological replicates. For proteins detected in fewer than three replicates, all data available are shown as points. Dashed lines represent a linear fit to the approximate zero-order kinetics (labeled with the slope and 95% confidence interval). (B) Experimental design to test for proteins released by eggs upon fertilization as an explanation for protein loss. Released proteins will increase in the media fraction over time. (C) Time series for proteins detected in both the egg and supernatant or the egg alone. These data were used to classify whether proteins were lost by release rather than degradation (by direct evidence or annotation; see text). This class comprised all but two of the significantly decreasing proteins (D) besides the known targets shown in A. (D) Time series of protein loss plotted by their absolute abundance (log10 transformed) and classified by the mechanism of loss. Error is visualized as in A. SSX2IP-L and NPDC1 homologs (black) are putative new degradation targets (see text for full criteria). (E) EM images of the cortical granules (CG) on the egg cortex. After egg activation, cortical granules fuse with the outer membrane and expel their contents.