Significance
Whether residing in or invading the host, enterobacteria have to deal with host-related stress conditions. These stress factors also serve as sensory cues, informing bacteria that they are present inside the host. Here, we report that the PhoQ/PhoP two-component system, which was known to sense several host-related environmental changes, responds to osmotic upshift, another key stimulus associated with the host. This sensing is proposed to rely on a mechanism that detects changes in the physical properties of the membrane. Thus, a single enterobacterial kinase, PhoQ, senses a major part of host-associated stimuli. The PhoQ-mediated osmosensing increases bacterial fitness under hyperosmotic conditions found inside the host, and it is likely to play an important role in the regulation of virulence.
Keywords: enterobacteria, osmolarity, virulence, signal transduction, stress response
Abstract
The PhoQ/PhoP two-component system plays an essential role in the response of enterobacteria to the environment of their mammalian hosts. It is known to sense several stimuli that are potentially associated with the host, including extracellular magnesium limitation, low pH, and the presence of cationic antimicrobial peptides. Here, we show that the PhoQ/PhoP two-component systems of Escherichia coli and Salmonella can also perceive an osmotic upshift, another key stimulus to which bacteria become exposed within the host. In contrast to most previously established stimuli of PhoQ, the detection of osmotic upshift does not require its periplasmic sensor domain. Instead, we show that the activity of PhoQ is affected by the length of the transmembrane (TM) helix as well as by membrane lateral pressure. We therefore propose that osmosensing relies on a conformational change within the TM domain of PhoQ induced by a perturbation in cell membrane thickness and lateral pressure under hyperosmotic conditions. Furthermore, the response mediated by the PhoQ/PhoP two-component system was found to improve bacterial growth recovery under hyperosmotic stress, partly through stabilization of the sigma factor RpoS. Our findings directly link the PhoQ/PhoP two-component system to bacterial osmosensing, suggesting that this system can mediate a concerted response to most of the established host-related cues.
The two-component systems are widely employed by bacteria to sense and respond to environmental changes (1). A prototypical two-component system consists of a membrane-imbedded sensor kinase and a cytosolic response regulator. When the sensor kinase is activated by an environmental stimulus, it autophosphorylates and then transfers the phosphate group to its cognate response regulator (2). The activated response regulator subsequently binds to specific gene promoters to regulate their expression, thus initiating cellular responses to environmental changes (3).
Out of about 30 two-component systems present in Escherichia coli or Salmonella species (1), the PhoQ/PhoP system is the major sensor for various host-associated environmental cues, regulating magnesium homeostasis, cell envelope composition, stress resistance, and virulence. It is known to respond to such stimuli as low divalent cations (4, 5), low pH (6), and the presence of cationic antimicrobial peptides (AMPs) (7). PhoQ, the dimeric sensor kinase of this system, consists of five domains, including the periplasmic sensor domain, the transmembrane (TM) domain, and three cytosolic domains: the HAMP domain involved in signal transmission, the dHp domain required for dimerization, and the catalytic domain. The dimeric sensor domain is reported to adopt different conformations when any of the three types of stimuli mentioned above is present (4–7). The sensing mechanisms for low magnesium and AMPs partially overlap, involving an acidic surface facing the membrane in the periplasmic sensor domain. Mutations in this region prevent PhoQ interactions with Mg2+ or AMPs and abrogate the response (7–9). Sensing of mild acidic pH involves both the sensor domain and the cytoplasmic domains (10, 11), with the latter detecting the cytoplasmic pH change. The PhoQ TM domain is a dimer of two TM helices that form a four-helix bundle (12). Upon activation of the sensory domain, the periplasmic ends of the two TM1 helices move closer to each other and push the two TM2 helices farther apart, accompanied by a TM2 rotation, which requires the solvation of a semichannel in the four-helix bundle (12). Asn202, located near the center of the TM2 helix and proximal to this semichannel, affects the solvation of the cavity, and mutations of Asn202 to a nonpolar residue lock PhoQ in the kinase-inactive state (13). Thus, a combination of scissoring and rotation of the helices was proposed as the mechanism of PhoQ activation, mediating signal transmission through the TM domain (12–14).
The phosphorylated response regulator PhoP activates the expression of a set of genes, which varies among different bacteria (15, 16). E. coli and Salmonella share several conserved ancestral genes of the PhoP regulon, including a magnesium transporter (mgtA in E. coli), the phoPQ operon itself, a negative regulator of PhoQ/PhoP (mgrB), and several other genes (17, 18). Notably, unlike most other genes in the PhoP regulon, the expression of mgtA in Salmonella was shown to be controlled by two additional regulatory factors: a Mg2+-binding riboswitch in the 5′-UTR of mgtA (19) and a proline-rich leader peptide MgtL, which may up-regulate mgtA transcription under proline limitation, such as that resulting from osmotic stress (20). A virulence gene mgtC in the PhoP regulon of Salmonella Typhimurium also encodes a proline-rich leader peptide in its 5′-UTR, which reportedly senses the proline level in vivo and promotes the translation of mgtC under hyperosmotic stress (21). However, another study suggested that mgtL translation might not respond to cytoplasmic proline level (22). Thus, the effects of osmolarity on the PhoP regulon remained unclear, despite the potential importance of hyperosmotic stress for the induction of virulence genes as a cue that E. coli or Salmonella encounters when entering the eukaryotic host.
In this study, we demonstrate that hyperosmotic stress is sensed by the PhoQ/PhoP system. We propose that the underlying osmosensing by PhoQ relies on a mechanism, whereby changes in the membrane properties under hyperosmotic conditions are sensed via conformational rearrangement within the TM domain of PhoQ. Finally, we show that the PhoQ/PhoP-mediated response to high osmolarity accelerates the recovery of bacterial cells from osmotic stress, which is partly due to the induction of σS-mediated stress response.
Results
PhoQ/PhoP System Is Activated by Osmotic Upshift.
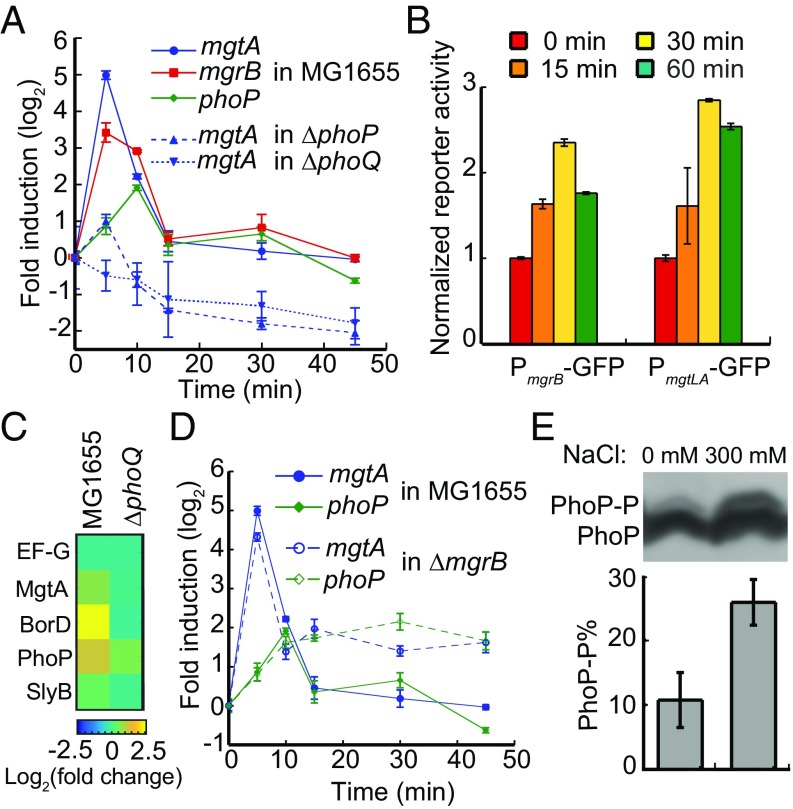
When testing transcription of E. coli PhoQ/PhoP-regulated genes upon stimulation with 300 mM NaCl, we observed a drastic (32-fold) but short-lived (15 min) up-regulation of mgtA (Fig. 1A). Other genes in the PhoP regulon, including mgrB, phoP, slyB, and borD, showed similar, but less prominent, transient transcriptional activation (SI Appendix, Fig. S1A). The maximum transcriptional up-regulation occurred around 5–10 min after such hyperosmotic stimulation, followed by an adaptation phase, when the transcription returned back to the prestimulation level. Since the EnvZ/OmpR two-component system is known to sense osmotic changes of the environment, we next investigated whether a cross-regulation from EnvZ/OmpR might be involved in the up-regulation of PhoP regulon during osmotic upshift. Among four single deletion strains (ΔphoQ, ΔphoP, ΔenvZ, and ΔompR), deletions of either envZ or ompR did not appear to affect the osmoresponses (SI Appendix, Fig. S1B), whereas deletions of either phoP or phoQ abrogated the responses almost entirely (Fig. 1A). Moreover, the time course of the OmpR regulon activation (SI Appendix, Fig. S1C) was clearly different from that of the PhoP regulon. Taken together, these results confirm that the observed osmoresponse requires PhoQ/PhoP, and it is different from and independent of the response mediated by the EnvZ/OmpR osmosensing system.
Fig. 1.
PhoQ senses osmotic upshift. (A) Changes in transcription of indicated PhoP-regulated genes after stimulation of exponentially growing E. coli wild-type, ΔphoQ, and ΔphoP strains with 300 mM NaCl at time point 0 (corresponding to OD600 = 0.4; see Materials and Methods for details). Transcription was monitored by RT-qPCR using primers within the coding regions of respective genes (SI Appendix, Table S2). The up-regulation of transcription is significant for mgtA, mgrB, and phoP in the wild-type cells (P < 0.001 according to t test), whereas it is not significant in ΔphoQ and ΔphoP strains (P > 0.3). (B) Expression of GFP reporter under control of mgrB or mgtLA promoters upon an osmotic upshift as in A. The values are normalized to the fluorescent signal at time point 0. The increase of GFP expression is significant at P < 0.05. (C) The heatmap of changes in amounts of indicated proteins after stimulation with 300 mM NaCl for 30 min in E. coli wild-type and ΔphoQ strains. Levels of all PhoP-regulated proteins increased significantly in the wild type (see SI Appendix, Table S3 for details). The amount of elongation factor G (EF-G), which was not affected by phoQ deletion, was used as a control. (D) Changes in transcription of mgtA and phoP in E. coli wild-type or ΔmgrB strains upon an osmotic upshift as in A. The data for mgtA and phoP transcription in the wild type are the same as in A. The up-regulation of transcription in both strains is significant at P < 0.001. (E) Phosphorylation of PhoP upon hyperosmotic treatment. E. coli ΔmgrB strain expressing PhoP-HA from a plasmid was grown with 0.006% arabinose to OD600 = 0.4. Then cells were either treated with 300 mM NaCl or left untreated for 2 more hours. Equal amounts of cells were then harvested and the phosphorylated PhoP (PhoP-P) in cell lysates was separated from the unphosphorylated species by phos-tag gel electrophoresis and visualized using immunoblotting with anti-HA primary antibody. The percentage of PhoP-P was quantified and presented as a bar chart Below. All data points represent averages of at least three independent experiments and error bars show SDs.
To further confirm that this transient transcriptional activation can lead to increased protein levels, we constructed reporter plasmids in which gfp was placed under control of the promoter regions of mgtLA (PmgtLA-GFP) or mgrB (PmgrB-GFP). When cells with these reporter plasmids were treated with 300 mM NaCl, we indeed observed a significant increase in GFP fluorescence measured by flow cytometry (Fig. 1B), although the maximal relative fluorescence increase was weaker and delayed compared with the transcriptional activation. These differences are expected, given the transient nature of gene activation and time needed for production, folding, maturation, and degradation of GFP proteins (23). Additionally, we performed proteomic analysis before and after NaCl treatment (Fig. 1C). We observed a pronounced increase in protein levels for MgtA, PhoP, BorD, and SlyB in the wild type, whereas no or little increase occurred in ∆phoQ cells. This further supports the physiological relevance of the observed activation of the PhoQ/PhoP two-component system by osmotic upshift.
The adaptation phase observed for the PhoQ/PhoP-dependent response to osmotic upshift indicates the involvement of negative feedback. A likely candidate for such a feedback regulator is a membrane peptide MgrB, which is transcriptionally activated by PhoP and interacts with PhoQ to inhibit its activity (24). Indeed, although no adaptation was originally observed for the PhoQ/PhoP-mediated response to magnesium limitation (24), a more recent study showed at least partial MgrB-dependent adaptation (25). We observed that adaptation to osmotic upshift was either entirely (for phoP) or partially (for mgtA) lost when mgrB was deleted (Fig. 1D), consistent with the proposed role of MgrB as a negative feedback regulator. Only partial loss of adaptation in the case of mgtA indicates that this gene might be under control of an additional regulatory factor. The mechanism of this additional regulation remains to be elucidated, but it appears to act faster than the one mediated by MgrB, which likely explains the difference in the time course of transcriptional activation between mgtA and phoP (Fig. 1 A and C).
Finally, to directly confirm the increased PhoQ-dependent phosphorylation of PhoP upon osmotic upshift, the level of phosphorylated PhoP (PhoP-P) was followed in vivo using phos-tag gels combined with immunoblotting. Because of the relatively low sensitivity of this approach (26) and the repressive growth condition for the native phoPQ operon (Materials and Methods) (4), PhoP could not be detected in the wild-type strain. Therefore, we expressed PhoP tagged at the C terminus with the human influenza hemagglutinin (PhoP-HA) from a plasmid, under induction with a low level of arabinose (0.006%). Furthermore, the experiments were performed in the ΔmgrB strain that shows persistent osmotic response. Under these conditions, we observed a clear increase in the level of PhoP-P after cells were treated with 300 mM NaCl for 2 h (Fig. 1D). These results further support our conclusions that the PhoQ/PhoP system is activated by osmotic upshift.
Because the PhoQ/PhoP two-component system is best studied in Salmonella, we also tested whether S. Typhimurium PhoQ, produced in the E. coli ΔphoQ strain from a plasmid induced by 0.002% arabinose, could activate the E. coli PhoP regulon upon an osmotic upshift. Indeed, the activation of mgtA transcription mediated by S. Typhimurium (ST) PhoQ was similar to the response mediated by similarly expressed E. coli (EC) PhoQ (SI Appendix, Fig. S2A). Notably, the transcriptional response in the ΔphoQ/EC_PhoQ strain was nearly indistinguishable from the wild-type response (Fig. 1A). Furthermore, after NaCl treatment the distribution of the induced PmgtLA-GFP reporter levels was only slightly broader in the ΔphoQ/EC_PhoQ strain compared with the wild type (SI Appendix, Fig. S2B). Both these results suggest that no significant artifacts are introduced by phoQ expression from a plasmid.
Characterization of PhoQ-Mediated Osmosensing.
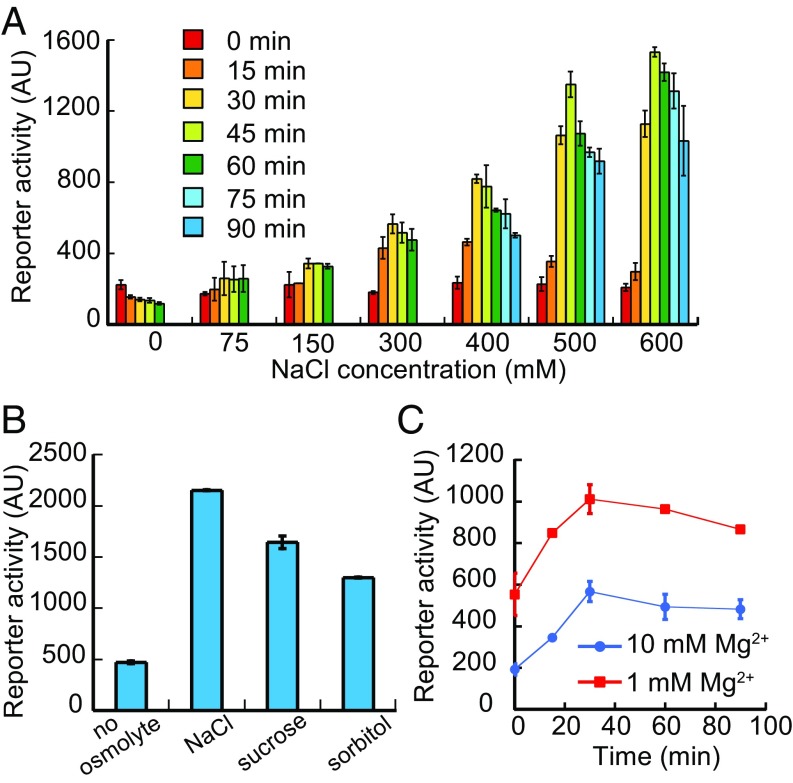
We next characterized the range of NaCl concentrations that can be sensed by PhoQ in E. coli, using the PmgtLA-GFP reporter (Fig. 2A). The response becomes significant above 150 mM NaCl and gradually reaches a plateau at NaCl concentrations of 500–600 mM. At all levels of stimulation the reporter activation peaked at 30–45 min and decayed afterward, consistent with the adaptive nature of the osmotic response. Besides ionic osmolytes, we observed that PhoQ/PhoP also responds to nonionic osmolytes, such as sucrose and sorbitol (Fig. 2B). The responses to the addition of 600 mM sucrose or 600 mM sorbitol were only marginally weaker than to the equivalent ionic stimulus, 300 mM NaCl. Notably, in these latter experiments the nonadapting ΔmgrB strain was used to facilitate measurements and interpretation of the data.
Fig. 2.
Characterization of PhoQ-mediated osmosensing. (A) PhoQ activity monitored via the expression of PmgtLA-GFP reporter in E. coli wild-type cells after stimulation with indicated NaCl concentrations. The P values (according to t test) of activation compared with the controls (0 min) are 0.3, 0.05, and 0.002 for stimulations with 75 mM, 150 mM, and 300 mM NaCl, respectively, and less than 0.001 for stimulations with 400 mM, 500 mM, and 600 mM NaCl. (B) PhoQ activity monitored via the expression of PmgtLA-GFP reporter in ∆mgrB strain after at least a 30-min incubation without or with indicated ionic (300 mM NaCl) or nonionic (600 mM sucrose, 600 mM sorbitol) osmolytes. The activations are significant at P < 0.01. (C) Activation of PmgtLA-GFP reporter after osmotic upshift (addition of 300 mM NaCl at time 0) in the presence of 10 mM or 1 mM magnesium in the growth media. The activation is significant at P < 0.01. All data points represent the averages of at least three independent experiments and error bars show SDs.
We also investigated mutual interplay between osmotic response and sensing of Mg2+ limitation. Other than the basal activity, a similar response to 300 mM NaCl was observed in the presence of 10 mM Mg2+ or 1 mM Mg2+ (Fig. 2C), suggesting that PhoQ is activated by an osmotic upshift independently of magnesium concentration. Notably, the magnitudes of osmotic response and activation by low magnesium were comparable. This additivity of responses contrasts to the activation of PhoQ by AMPs, which was previously shown to occur at low but not at high levels of Mg2+ (10 mM) (7), further suggesting that sensing of osmolarity and magnesium limitation rely on different mechanisms.
The Mechanism of PhoQ Osmosensing.
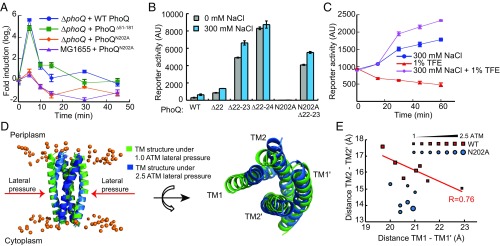
The previously established PhoQ stimuli—the limitation of external divalent cations, acidic pH, and cationic AMPs (4, 6, 7)—are sensed solely or at least partially via the periplasmic sensor domain of PhoQ. To test whether the sensor domain is also involved in detection of hyperosmotic stimuli, we constructed a corresponding PhoQ deletion mutant (PhoQΔ51–181). Surprisingly, cells expressing this mutant exhibited comparable induction of mgtA transcription (Fig. 3A) and PmgtLA-GFP reporter expression (SI Appendix, Fig. S2C) as cells expressing the wild-type PhoQ, which demonstrates that the sensor domain of PhoQ is not essential for osmosensing.
Fig. 3.
PhoQ-mediated osmosensing relies on the transmembrane domain. (A) Transcriptional response mediated by the wild-type PhoQ or mutants that lack the sensor domain (PhoQ∆51–181) or carry mutation in the TM domain (PhoQN202A), expressed from a plasmid at 0.002% arabinose induction in the ∆phoQ strain. Transcription of mgtA was monitored upon addition of 300 mM NaCl. The transcription up-regulation is significant for ΔphoQ strains complemented with the wild-type PhoQ or PhoQ∆51–181 (P < 0.005). (B) Activity of PmgtLA-GFP reporter in the ∆phoQ strains expressing either the wild type or PhoQ mutants. Cells grown to OD600 = 0.4 in the presence of 10 mM magnesium and 0.002% arabinose were stimulated with 300 mM NaCl for 30 min, and their fluorescence was measured before (labeled as 0 mM NaCl) and after the stimulation (labeled as 300 mM NaCl). The P values for the activation of the wild type and PhoQ mutants (PhoQΔ22, PhoQΔ22–23, PhoQΔ22–24, and PhoQΔ22–23+N202A) are 0.002, 0.02, 0.05, 0.08, and 0.02, respectively. Normalized data can be found in SI Appendix, Fig. S3A for clear visualization of error bars. (C) Time course of PmgtLA-GFP reporter activation in ∆mgrB cells upon stimulation with 300 mM NaCl, 1% TFE, or both stimuli combined, as indicated. The activation is significant with P < 0.01 for stimulation with 300 mM NaCl or both 1% TFE and 300 mM NaCl. (D) Simulated structures of the wild-type PhoQ TM helices in the 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) membrane bilayer at 1.0 (green) and 2.5 (blue) atm lateral pressure (see Materials and Methods and SI Appendix, SI Materials and Methods for details of simulations), representing side (Left image) and top (Right image) views. The phosphorus atoms of POPE are represented as orange balls to indicate the interface between solvent and the membrane. (E) Conformational changes in the TM domain of the wild type and PhoQN202A under indicated lateral membrane pressure, represented as the distance between termini of helices at the periplasmic surface. The P values for TM domain conformation change are 0.69 and less than 0.05 for PhoQN202A and the wild type, respectively. Data points in A–C represent the averages of at least three independent experiments and error bars show SDs.
One prominent morphological change under hyperosmotic shock is the reduction of the cell volume due to the loss of water (27). This causes several physical changes in the membrane, including increases in thickness, curvature, and lateral pressure. The increase of membrane thickness leads to a temporary hydrophobic mismatch (28), i.e., the hydrophobic thickness of the lipid bilayer in the immediate vicinity of PhoQ becomes larger than the hydrophobic length of the TM domain. This mismatch creates a hydrophilic/hydrophobic interface, thus resulting in an energetically unfavorable state (28). Relaxation of this state requires either deformation of the lipid bilayer in the vicinity of PhoQ or the conformational change within the TM domain. Given that the conformation of the transmembrane domain is reportedly important for activation of PhoQ (12–14), we hypothesized that PhoQ activation by an osmotic upshift may result from a conformational change that rearranges transmembrane helices to minimize the hydrophobic mismatch.
To test this hypothesis, we mimicked the hydrophobic mismatch by making three deletions within TM helix 1, which decreased the helix length (PhoQΔ22, PhoQΔ22–23, and PhoQΔ22–24). These sites were chosen as little or no cross-linking with other TM residues was observed at these three positions in a previous study (12). We observed that these deletions progressively elevated basal activity of PhoQ (Fig. 3B), from about 4-fold to 40-fold, with PhoQΔ22–24 having the strongest effect. The deletion mutants responded to a hyperosmotic stimulus in degrees that were inversely correlated with the basal activities of individual mutants (Fig. 3B and SI Appendix, Fig. S3A). Interestingly, all three mutants completely lost their sensitivity to magnesium (SI Appendix, Fig. S3B), suggesting that TM helix 1 shortening may prevent signal transduction from the sensor domain to the transmembrane domain.
We also tested the osmoresponse of the previously reported N202A TM substitution mutant (PhoQN202A), which does not respond to magnesium limitation due to the mutation locking PhoQ in a kinase-inactive state (13). We observed that PhoQN202A was similarly unresponsive to an osmotic upshift (Fig. 3 A and B). Nevertheless, N202 is not required for osmosensing, as a double mutant (PhoQN202A+Δ22–23) with both N202A and TM shortening mutations showed elevated basal kinase activities and significant responses to an osmotic upshift (Fig. 3B).
To further examine the potential role of membrane lateral pressure in PhoQ osmosensing, we used 2,2,2-trifluoroethanol (TFE), a small chemical that is known to insert into membrane and increase lateral pressure in the head-group region while decreasing it in the middle section of the lipid bilayer (29). Although no increase of PhoQ activity was observed in ∆mgrB cells when TFE was added alone, TFE could apparently potentiate the effect of the osmotic upshift caused by 300 mM NaCl (Fig. 3C). These results imply that changes of membrane lateral pressure by TFE insertion further activate PhoQ, even though these changes alone are not enough to switch PhoQ from the kinase-inactive to the kinase-active state.
To verify the proposed mechanism of PhoQ activation, we performed long-timescale explicit solvent molecular dynamics (MD) simulations of the PhoQ transmembrane domain under increased lateral pressure across the membrane (Fig. 3D). Here we relied on a previously published MD model that was refined by disulfide cross-linking data (12) and applied increased lateral pressure to mimic the effect of osmolarity on cell membrane (30, 31). Notably, in our simulations the increase in the lateral membrane pressure also increases thickness of the lipid bilayer, thus inherently relating these two hyperosmolarity-induced perturbations. Our data suggest that increased lateral membrane pressure induces a rearrangement within the transmembrane domain of the wild-type PhoQ, pushing closer the ends of TM1 helices and forcing the ends of TM2 helices further apart (Fig. 3 D and E). The conformational change observed in our simulations agrees with the published active conformation of PhoQ (12). As a negative control, no significant conformational changes were observed for PhoQN202A, consistent with the experimentally observed lack of response in this mutant (Fig. 3E). Cumulatively, our results strongly support the hypothesis that PhoQ senses osmotic upshift through conformational changes caused by interactions of its TM domain with the osmotically perturbed membrane.
PhoQ/PhoP Enhances Growth Recovery from Hyperosmotic Stress.
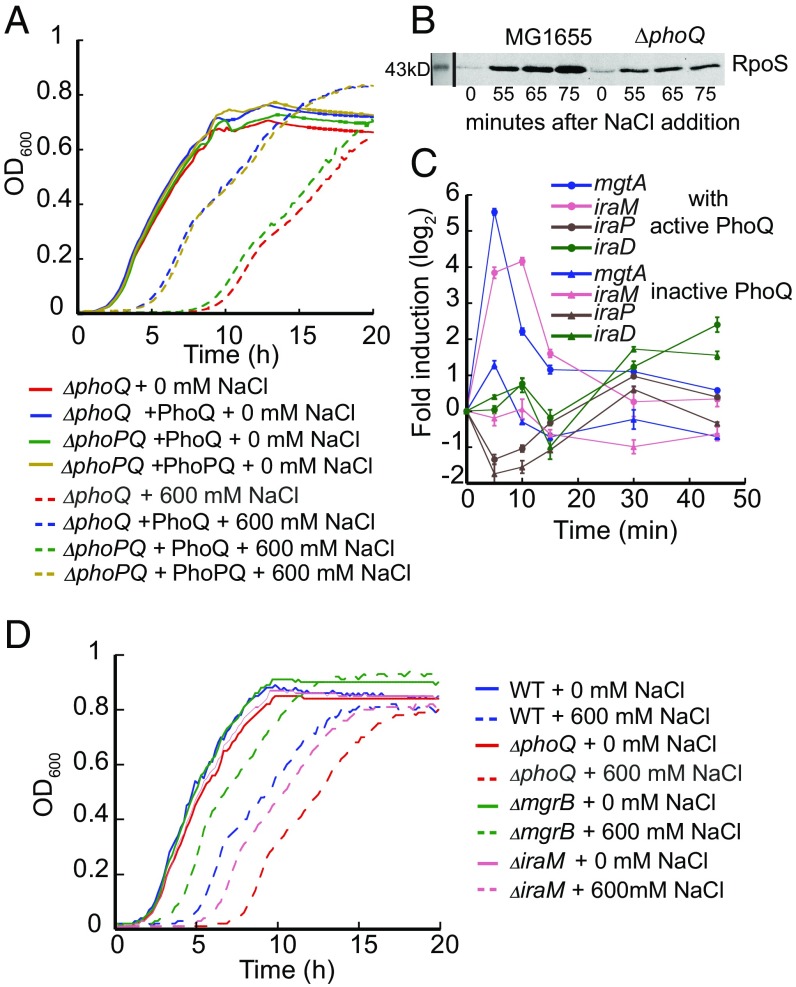
To investigate whether activation of the PhoQ/PhoP system might directly benefit bacterial cells under osmotic stress, we monitored growth of E. coli strains that lack PhoQ and/or PhoP upon culture dilution in media with varying osmolarity. While all strains tested displayed nearly identical growth without osmotic stress, the strains lacking either PhoP or PhoQ exhibited a significantly longer lag phase in the presence of osmotic stress than the wild-type cells (Fig. 4A and SI Appendix, Fig. S4). This effect could be complemented by expression of the missing proteins from the plasmid. As growth rates of all strains were similar in the logarithmical phase, PhoQ/PhoP-mediated osmotic response appears to specifically accelerate the resumption of growth at high osmolarity. Under our growth conditions, clear effects of osmotic stress on growth of the wild-type or mutant strains were observed starting from 500 mM NaCl, with the largest difference in growth recovery between the wild-type and ΔphoQ strains at around 600 mM NaCl (SI Appendix, Fig. S4A). Although 600 mM NaCl corresponds to higher osmolarity than that observed in mammalian blood and tissues, such osmolarity can be reached in the mammalian gastrointestinal tract (32, 33), meaning that it is physically relevant to enterobacteria.
Fig. 4.
PhoQ mediates resistance to hyperosmotic conditions partly by stabilizing RpoS (σS). (A) Growth of indicated E. coli strains in the absence or presence of 600 mM NaCl. PhoQ or PhoP/PhoQ (PhoPQ) were expressed from respective plasmids. Because deletion of phoP has a polar effect on the expression of phoQ, a ΔphoPphoQ strain complemented with PhoQ was used to elucidate the specific effect of phoP. The growth curves are representatives of at least three independent experiments. (B) The amount of σS in the wild-type and ∆phoQ strains at indicated time points after 600 mM NaCl addition, monitored via immunoblotting with anti-RpoS primary antibody. Equal amounts of cells were collected at each time point for cell lysate preparation and equal amounts of cell lysate were loaded in each lane. The immunoblot is a representative of three independent experiments. (C) Transcription profiles of mgtA and of σS anti-adaptor genes (iraM, iraP, and iraD), measured by RT-qPCR after addition of 300 mM NaCl to E. coli expressing the wild-type PhoQ or PhoQN202A. All data points represent the averages of three independent experiments and error bars show SDs. The transcription up-regulation is significant at P < 0.0001 for mgtA and iraM in E. coli expressing the wild-type PhoQ. (D) Growth of indicated E. coli strains in the absence or presence of 600 mM NaCl, as in A.
We attempted to further identify PhoP target genes that may be responsible for E. coli growth recovery under osmotic stress. We first made deletion of several genes in the PhoP regulon that show high or moderate induction upon hyperosmotic stimulation (mgtA, borD, and yrbL). However, all these single-deletion strains behaved similarly to the wild type (SI Appendix, Fig. S4B), indicating that these genes might not be individually important for the osmotic stress resistance or play only a minor role. We next explored the possibility that the observed stress resistance was achieved via the alternative sigma factor σS (RpoS). Stabilization of σS is known to be promoted by several anti-adaptor proteins, including a PhoQ/PhoP-regulated protein IraM (IraP in Salmonella) (34–36). Sigma factor σS mediates general stress response when cells face various environmental stress conditions, including osmotic upshift (37, 38), and increase of σS might potentially explain the PhoQ/PhoP-dependent acceleration of the growth recovery under stress. Therefore, we examined the amount of σS in the wild-type and ΔphoQ strains as well as the induction of iraM under osmotic stress. Indeed, the amount of σS steadily increases in E. coli wild-type cells during exposure to the hyperosmotic stress, whereas the PhoQ deletion strain shows only a weak initial elevation of σS levels within the time window examined (Fig. 4B). In parallel, E. coli cells expressing the wild-type PhoQ showed about 20-fold induction of iraM transcription after the osmotic upshift, whereas those carrying inactive PhoQN202A showed no activation (Fig. 4C). Two other known anti-adaptors in E. coli, IraP and IraD, did not display pronounced changes in transcription under osmotic stress (Fig. 4C). Therefore, we hypothesized that under hyperosmotic conditions, the PhoQ/PhoP activates the anti-adaptor protein IraM, which stabilizes the σS by preventing its degradation, leading to faster growth recovery.
To confirm this mechanism, we constructed the ΔiraM strain and tested it in growth experiments. While having a similar growth rate in the logarithmical phase, the ΔiraM strain displayed a longer lag phase than the wild type (Fig. 4D). However, it recovered noticeably faster than the ΔphoQ strain, suggesting that IraM-mediated stabilization of σS can only partially explain the PhoQ/PhoP-mediated resistance to osmotic stress, and further studies are needed to fully elucidate the underlying regulation. We also observed that the ΔmgrB strain exhibited an even shorter lag phase at high osmolarity compared with the wild type (Fig. 4D), consistent with the higher activity of the PhoQ/PhoP system in the absence of MgrB.
Discussion
The PhoQ/PhoP two-component system is well known to respond to several host-associated cues, such as the levels of magnesium, pH, and AMPs (4–7). In contrast, high osmolarity, another major stimulus encountered by enterobacteria within the host (39), was up to now only suggested to affect translation of individual PhoP-regulated genes (20, 21). In this study, we demonstrate that E. coli and Salmonella PhoQ is activated by osmotic upshift.
The magnitude of the observed hyperosmotic stimulation of the PhoQ/PhoP system was comparable to its activation by low magnesium or by other established stimuli (6, 7, 11, 13, 24). However, in contrast to all other established stimuli (except cytoplasmic pH) the activation of PhoQ by osmotic upshift does not require the sensor domain but depends on the transmembrane region. Our data suggest that PhoQ senses hyperosmotic stimuli through the interaction of its transmembrane domain with the lipid bilayer, apparently responding to its increased thickness and lateral pressure under hyperosmotic stress. We propose that the resulting hydrophobic mismatch between the thickness of the bilayer and the height of the transmembrane domain is the primary signal for PhoQ. Similar changes in physical properties of membranes are known to regulate membrane-spanning channels and transporters (40, 41) as well as the thermosensor DesK in Bacillus subtilis (42). Notably, the mechanism of osmosensing by PhoQ is distinctly different from that of EnvZ or CpxA, other two-component kinases that are known to mediate osmoresponse in E. coli and Salmonella. EnvZ senses both hypoosmotic and hyperosmotic stimuli via a conformational change of its cytoplasmic domain (43), whereas CpxA senses cell envelope stresses via an unknown mechanism (44). Nevertheless, osmoresponses mediated by EnvZ/OmpR and PhoQ/PhoP might be connected, as it has been reported that ProP, an OmpR-regulated protein, is also indirectly controlled by PhoQ/PhoP in E. coli (45).
Interestingly, we observed that the osmotic response mediated by PhoQ/PhoP has a more pronounced adaptation phase than responses to other stimuli (24, 25). This adaptation phase appears to be mediated by at least two types of negative feedback. One depends on MgrB, a membrane protein known to be a part of the PhoP regulon and to negatively regulate PhoQ (24). Why the MgrB-mediated adaptation might be stronger in the osmotic response compared with other PhoQ-mediated responses remains unknown. But it is possible that besides up-regulation of MgrB expression, the membrane perturbation caused by osmotic upshift may additionally enhance the MgrB/PhoQ interaction. An additional fast feedback, revealed in the absence of MgrB, appears to regulate specifically mgtA. The nature of this feedback remains to be elucidated in further studies. The activation of mgtA transcription was also greater than for other tested PhoP-regulated genes. This may be due to the higher affinity of the mgtA promoter to PhoP (18) and/or to the additional osmolarity-dependent regulation provided by the leader region of mgtA (20).
In Salmonella, osmolarity is known to be one of the environmental factors that promote invasion and virulence (46, 47), but details of this regulation have not been understood. Moreover, while PhoQ/PhoP is essential for Salmonella virulence in phagocytes, it seems to have complex effects on invasion and it is only one of the relevant regulatory systems. On one hand, the activated PhoQ/PhoP indirectly represses the expression of the type III secretion system, which is the major factor for invasion of epithelial cells (48). On the other hand, activation of the PhoQ/PhoP system induces other virulence genes, such as pagN coding for an outer membrane protein that promotes adhesion and invasion of host cells (49, 50). Interestingly, overexpression of pagN partially compensates SPI-1 deletion (49), suggesting Salmonella encodes multiple pathways for host cell invasion and PhoQ/PhoP is involved in regulation of this process. In addition, our results further suggest that the PhoQ/PhoP-mediated osmosensing may confer an immediate fitness benefit in the host environment, by accelerating resumption of growth under hyperosmotic stress.
In summary, our work suggests that PhoQ integrates a range of host-related cues due to its versatile sensory capabilities (SI Appendix, Fig. S5). Additionally, the PhoQ/PhoP system can be indirectly activated by other host-associated inputs, because MgrB inhibition of PhoQ is alleviated in a reduced environment (51) and another small regulator protein SafA connects it to the acid-resistance system (52). All of these further emphasize the importance of the PhoQ/PhoP system for enterobacteria to switch lifestyle inside the host.
Materials and Methods
Strains and Plasmids.
The E. coli strains and plasmids used in this study are listed in SI Appendix, Table S1. All E. coli strains were derived from the parental strain MG1655.
Bacterial Growth.
E. coli strains were grown in Luria–Bertani (LB) medium overnight at 37 °C, then diluted 1:100 to fresh medium A (7 g of nutrient broth, 1 g of yeast extract, 2 g of glycerol, 3.7 g of K2HPO4, and 1.3 g of KH2PO4 per liter) (53), supplemented with 10 mM MgSO4 or as indicated otherwise. Where appropriate, 50 μg/mL kanamycin, 34 μg/mL chloramphenicol, or 100 μg/mL ampicillin was added. The cultures were subsequently grown to early log phase (OD600 = 0.4) at 37 °C. Where indicated, at this point various osmolytes were added and aliquots of cultures were collected before and after the addition at different time points. When indicated, 0.002% or 0.006% l-arabinose was added at the start of the day culture to induce protein expression.
For growth curve analysis, E. coli strains were grown to early log phase (OD600 = 0.4) in medium A, where appropriate, with antibiotics and arabinose, and then diluted 1:100 to the same media with or without NaCl addition. The growth of the strains was monitored with an Infinite M1000 microplate reader (Tecan).
Total RNA Extraction and RT-qPCR.
Cell pellets collected at different time points after hyperosmotic treatment were resuspended and lysed by homogenization using a Precellys evolution homogenizer (Bertin), followed by total RNA purification using the mirVana miRNA isolation kit (Ambion). Purified total RNA was then used as a template (0.2 ng/µL) for RT-qPCR analysis using the KAPA SYBR fast universal one-step qRT-PCR kit (Kapa Biosystems) following the manufacturer’s instructions. The gene ssrA, coding for tmRNA, was used as internal control. The primers used for qPCR analysis and their locations are listed in SI Appendix, Table S2. The RT-qPCR was performed using a CFX384 Touch real-time PCR detection system (Bio-Rad). Each sample was tested in triplicates and data were analyzed using CFX Manager software (Bio-Rad).
Construction and Expression of GFP Reporter Genes.
The intergenic region upstream of mgtLA or mgrB was cloned between the XhoI and BamHI sites and fused to a GFP gene in pUA66 plasmid, and the resulting plasmids were then transformed to strains as indicated. To test the expression of the GFP reporter gene, transformed cells were grown overnight and then diluted 1:100 to fresh medium A supplemented with magnesium as indicated. The cultures were treated with osmolytes or TFE at OD600 = 0.4 and the fluorescence of cells at different time points was monitored with a BD LSRFortessa SORP flow cytometer (BD Biosciences), equipped with a 100-mW 488-nm laser and a 510/20 bandpass filter. The acquired data were analyzed using BD FACSDiva software version 8.0 (BD Biosciences).
Proteomic Analysis.
E. coli wild-type and ΔphoQ strains were grown to OD600 = 0.4 in medium A supplemented with 10 mM MgSO4, followed by addition of NaCl to a final concentration of 300 mM. Equal amounts of cells were collected before and 30 min after the NaCl addition. Cell pellets were washed three times with ice-cold PBS and subjected to proteomic analysis using mass spectrometry (MS). Details of proteomic analysis can be found in SI Appendix, SI Materials and Methods.
Phos-Tag Gel Analysis.
Cells were grown in medium A supplemented with 10 mM MgSO4 and 0.006% arabinose to OD600 = 0.4 and divided into two equal portions, one of which was treated with 300 mM NaCl while the other was left untreated. After 2 h, equal amounts of cells were harvested for each growth condition, and cell pellets were washed with ice-cold 10 mM Tris-Cl (pH 6.8) buffer and flash frozen in liquid nitrogen. To prepare cell lysate, cell pellets were resuspended in 90 μL buffer (containing 10 mM Tris-Cl, pH 6.8 and EDTA-free proteinase inhibitor) on ice, lysed by homogenization, and followed by centrifugation to clear up the lysate at 4 °C.
Cell lysates (45 μL) combined with 15 μL of 4× SDS loading buffer were mixed vigorously for 30 s at room temperature and 10 μL of the mixture was loaded onto a phos-tag gel (8% 19:1 polyacrylamide gel with 75 μM phos-tag). Proteins were separated by electrophoresis at 4 °C and transferred to a PVDF membrane in transfer buffer containing 20% methanol and 0.05% SDS. PhoP-HA was detected with anti-HA primary antibody (Sigma) and IRDye 800CW-conjugated secondary antibody (LI-COR). The protein bands were visualized with the Odyssey CLx imaging system (LI-COR) and quantified with ImageJ.
MD Simulations.
Explicit solvent MD simulations were performed using a previously constructed model of the PhoQ transmembrane dimer (12) consisting of TM1 (15VRFLLATAAVVLVLSLAYGMVALIGYSVSFDKT47) and TM2 (190MVWSWFIYVLSANLLLVIPLLWVAAWWSLRP220) helices. All of the simulations were conducted using the GROMACS 5.1.2 software package (54) with CHARMM36 force field (55) and TIP3P (55) water model under particle mesh Ewald cubic periodic boundary conditions. Simulation details can be found in SI Appendix.
Analysis of RpoS Levels by Immunoblotting.
Cell lysates were prepared as described before with modifications (56). Specifically, cells were grown to OD600 = 0.4 in medium A and treated with 600 mM NaCl. Samples were collected at different time points by pelleting equal amounts of cells and flash freezing pellets in liquid nitrogen. For the immunoblot analysis, cell pellets were resuspended in 55 μL 1× BugBuster protein extraction reagent (Novagen) supplemented with 0.1% lysonase (Novagen) and lysed by pipetting up and down for 10 s. The lysates were heated at 95 °C for 5 min after the addition of 18 μL 4× loading buffer and equal amount of lysates were loaded on a 10% SDS polyacrylamide gel. Proteins were separated by gel electrophoresis and transferred to a nitrocellulose membrane. Sigma factor RpoS was detected with specific primary antibody (BioLegend) and horseradish peroxidase-conjugated secondary antibody (Sigma). Protein bands were detected by chemiluminescence using an ADVANCED imager (Intas).
Supplementary Material
Acknowledgments
We thank Thomas Lemmin for providing PhoQ TM dimer coordinates, Jörg Kahnt for assisting the proteomic analysis, Frithjof Henkel for his excellent technical support, Abiola Pollard and Miriam Fischer for valuable discussions, and the LOEWE Center for Scientific Computing at the Goethe University Frankfurt for providing access to the high-performance computer clusters. This work was supported by Grant 294761-MicRobE from the European Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717272114/-/DCSupplemental.
References
- 1.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 2.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 3.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 4.García Véscovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: Environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 5.Véscovi EG, Ayala YM, Di Cera E, Groisman EA. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+ J Biol Chem. 1997;272:1440–1443. doi: 10.1074/jbc.272.3.1440. [DOI] [PubMed] [Google Scholar]
- 6.Prost LR, et al. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Bader MW, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 8.Cho US, et al. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J Mol Biol. 2006;356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 9.Waldburger CD, Sauer RT. Signal detection by the PhoQ sensor-transmitter. Characterization of the sensor domain and a response-impaired mutant that identifies ligand-binding determinants. J Biol Chem. 1996;271:26630–26636. doi: 10.1074/jbc.271.43.26630. [DOI] [PubMed] [Google Scholar]
- 10.Hicks KG, et al. Acidic pH and divalent cation sensing by PhoQ are dispensable for systemic salmonellae virulence. eLife. 2015;4:e06792. doi: 10.7554/eLife.06792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi J, Groisman EA. Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol Microbiol. 2016;101:1024–1038. doi: 10.1111/mmi.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemmin T, Soto CS, Clinthorne G, DeGrado WF, Dal Peraro M. Assembly of the transmembrane domain of E. coli PhoQ histidine kinase: Implications for signal transduction from molecular simulations. PLoS Comput Biol. 2013;9:e1002878. doi: 10.1371/journal.pcbi.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg SD, Clinthorne GD, Goulian M, DeGrado WF. Transmembrane polar interactions are required for signaling in the Escherichia coli sensor kinase PhoQ. Proc Natl Acad Sci USA. 2010;107:8141–8146. doi: 10.1073/pnas.1003166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molnar KS, et al. Cys-scanning disulfide crosslinking and bayesian modeling probe the transmembrane signaling mechanism of the histidine kinase, PhoQ. Structure. 2014;22:1239–1251. doi: 10.1016/j.str.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez JC, et al. Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet. 2009;5:e1000428. doi: 10.1371/journal.pgen.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. The promoter architectural landscape of the Salmonella PhoP regulon. Mol Microbiol. 2012;84:463–485. doi: 10.1111/j.1365-2958.2012.08036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato A, Tanabe H, Utsumi R. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: Identification of extracellular Mg2+-responsive promoters. J Bacteriol. 1999;181:5516–5520. doi: 10.1128/jb.181.17.5516-5520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minagawa S, et al. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J Bacteriol. 2003;185:3696–3702. doi: 10.1128/JB.185.13.3696-3702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 20.Park SY, Cromie MJ, Lee EJ, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee EJ, Choi J, Groisman EA. Control of a Salmonella virulence operon by proline-charged tRNAPro. Proc Natl Acad Sci USA. 2014;111:3140–3145. doi: 10.1073/pnas.1316209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao G, Kong W, Weatherspoon-Griffin N, Clark-Curtiss J, Shi Y. Mg2+ facilitates leader peptide translation to induce riboswitch-mediated transcription termination. EMBO J. 2011;30:1485–1496. doi: 10.1038/emboj.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingalls BP. Mathematical Modeling in Systems Biology: An Introduction. MIT Press; Cambridge, MA: 2013. [Google Scholar]
- 24.Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5:e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salazar ME, Podgornaia AI, Laub MT. The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics. Mol Microbiol. 2016;102:430–445. doi: 10.1111/mmi.13471. [DOI] [PubMed] [Google Scholar]
- 26.Park SY, Groisman EA. Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol Microbiol. 2014;91:135–144. doi: 10.1111/mmi.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood JM. Osmosensing by bacteria: Signals and membrane-based sensors. Microbiol Mol Biol Rev. 1999;63:230–262. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen OS, Koeppe RE., 2nd Bilayer thickness and membrane protein function: An energetic perspective. Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 29.van den Brink-van der Laan E, Chupin V, Killian JA, de Kruijff B. Stability of KcsA tetramer depends on membrane lateral pressure. Biochemistry. 2004;43:4240–4250. doi: 10.1021/bi036129d. [DOI] [PubMed] [Google Scholar]
- 30.Xie JY, Ding GH, Karttunen M. Molecular dynamics simulations of lipid membranes with lateral force: Rupture and dynamic properties. Biochim Biophys Acta. 2014;1838:994–1002. doi: 10.1016/j.bbamem.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Muddana HS, Gullapalli RR, Manias E, Butler PJ. Atomistic simulation of lipid and DiI dynamics in membrane bilayers under tension. Phys Chem Chem Phys. 2011;13:1368–1378. doi: 10.1039/c0cp00430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houpt TR, Houpt KA, Swan AA. Duodenal osmoconcentration and food intake in pigs after ingestion of hypertonic nutrients. Am J Physiol. 1983;245:R181–R189. doi: 10.1152/ajpregu.1983.245.2.R181. [DOI] [PubMed] [Google Scholar]
- 33.Hallbäck DA, Jodal M, Mannischeff M, Lundgren O. Tissue osmolality in intestinal villi of four mammals in vivo and in vitro. Acta Physiol Scand. 1991;143:271–277. doi: 10.1111/j.1748-1716.1991.tb09232.x. [DOI] [PubMed] [Google Scholar]
- 34.Hengge R. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res Microbiol. 2009;160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008;68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- 36.Schweder T, Lee KH, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landini P, Egli T, Wolf J, Lacour S. sigmaS, a major player in the response to environmental stresses in Escherichia coli: Role, regulation and mechanisms of promoter recognition. Environ Microbiol Rep. 2014;6:1–13. doi: 10.1111/1758-2229.12112. [DOI] [PubMed] [Google Scholar]
- 38.Hengge-Aronis R. Back to log phase: Sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 39.Fang FC, Frawley ER, Tapscott T, Vázquez-Torres A. Bacterial stress responses during host infection. Cell Host Microbe. 2016;20:133–143. doi: 10.1016/j.chom.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott JR, Needham D, Dilger JP, Haydon DA. The effects of bilayer thickness and tension on gramicidin single-channel lifetime. Biochim Biophys Acta. 1983;735:95–103. doi: 10.1016/0005-2736(83)90264-x. [DOI] [PubMed] [Google Scholar]
- 41.Mobashery N, Nielsen C, Andersen OS. The conformational preference of gramicidin channels is a function of lipid bilayer thickness. FEBS Lett. 1997;412:15–20. doi: 10.1016/s0014-5793(97)00709-6. [DOI] [PubMed] [Google Scholar]
- 42.Martín M, de Mendoza D. Regulation of Bacillus subtilis DesK thermosensor by lipids. Biochem J. 2013;451:269–275. doi: 10.1042/BJ20121825. [DOI] [PubMed] [Google Scholar]
- 43.Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raivio TL. Everything old is new again: An update on current research on the Cpx envelope stress response. Biochim Biophys Acta. 2014;1843:1529–1541. doi: 10.1016/j.bbamcr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 45.Eguchi Y, et al. Signal transduction cascade between EvgA/EvgS and PhoP/PhoQ two-component systems of Escherichia coli. J Bacteriol. 2004;186:3006–3014. doi: 10.1128/JB.186.10.3006-3014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altier C. Genetic and environmental control of salmonella invasion. J Microbiol. 2005;43:85–92. [PubMed] [Google Scholar]
- 47.Shen S, Fang FC. Integrated stress responses in Salmonella. Int J Food Microbiol. 2012;152:75–81. doi: 10.1016/j.ijfoodmicro.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 49.Lambert MA, Smith SG. The PagN protein of Salmonella enterica serovar Typhimurium is an adhesin and invasin. BMC Microbiol. 2008;8:142. doi: 10.1186/1471-2180-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert MA, Smith SG. The PagN protein mediates invasion via interaction with proteoglycan. FEMS Microbiol Lett. 2009;297:209–216. doi: 10.1111/j.1574-6968.2009.01666.x. [DOI] [PubMed] [Google Scholar]
- 51.Lippa AM, Goulian M. Perturbation of the oxidizing environment of the periplasm stimulates the PhoQ/PhoP system in Escherichia coli. J Bacteriol. 2012;194:1457–1463. doi: 10.1128/JB.06055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eguchi Y, Ishii E, Hata K, Utsumi R. Regulation of acid resistance by connectors of two-component signal transduction systems in Escherichia coli. J Bacteriol. 2011;193:1222–1228. doi: 10.1128/JB.01124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawaji H, Mizuno T, Mizushima S. Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins O-8 and O-9 of Escherichia coli K-12. J Bacteriol. 1979;140:843–847. doi: 10.1128/jb.140.3.843-847.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 55.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao R, Stock AM. Probing kinase and phosphatase activities of two-component systems in vivo with concentration-dependent phosphorylation profiling. Proc Natl Acad Sci USA. 2013;110:672–677. doi: 10.1073/pnas.1214587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.