Significance
Mycobacteria use ESX systems to transport protein substrates across the cytoplasmic membrane. The ESX-1 system is required for mycobacterial pathogenesis in Mycobacterium tuberculosis (M. tb), the cause of tuberculosis (TB). Differences in the expression of genes encoding ESX substrates directly impacts M. tb transmission and virulence. Deletion of genes encoding ESX exporters results in reduced levels of ESX substrates in mycobacteria. Here, we define a fundamental mechanism of regulation of ESX-1 substrates in M. marinum, a pathogenic mycobacterial species and a model for M. tb. We demonstrate that the transcriptional regulation of genes encoding ESX-1 substrates is linked to the presence or absence of the ESX-1 exporter. These findings provide insight into how substrate levels are intricately controlled in mycobacteria.
Keywords: regulation, protein secretion, ESX-1, ESAT-6, Mycobacterium
Abstract
ESX (ESAT-6 system) export systems play diverse roles across mycobacterial species. Interestingly, genetic disruption of ESX systems in different species does not result in an accumulation of protein substrates in the mycobacterial cell. However, the mechanisms underlying this observation are elusive. We hypothesized that the levels of ESX substrates were regulated by a feedback-control mechanism, linking the levels of substrates to the secretory status of ESX systems. To test this hypothesis, we used a combination of genetic, transcriptomic, and proteomic approaches to define export-dependent mechanisms regulating the levels of ESX-1 substrates in Mycobacterium marinum. WhiB6 is a transcription factor that regulates expression of genes encoding ESX-1 substrates. We found that, in the absence of the genes encoding conserved membrane components of the ESX-1 system, the expression of the whiB6 gene and genes encoding ESX-1 substrates were reduced. Accordingly, the levels of ESX-1 substrates were decreased, and WhiB6 was not detected in M. marinum strains lacking genes encoding ESX-1 components. We demonstrated that, in the absence of EccCb1, a conserved ESX-1 component, substrate gene expression was restored by constitutive, but not native, expression of the whiB6 gene. Finally, we found that the loss of WhiB6 resulted in a virulent M. marinum strain with reduced ESX-1 secretion. Together, our findings demonstrate that the levels of ESX-1 substrates in M. marinum are fine-tuned by negative feedback control, linking the expression of the whiB6 gene to the presence, not the functionality, of the ESX-1 membrane complex.
Bacteria use secretion systems to transport protein substrates across membranes. Gram-negative (or diderm-LPS) bacteria secrete protein across the inner membrane and outer membrane (OM) using type I–IX secretion systems (1). Protein secretion in Gram-negative bacteria is a tightly regulated process. In particular, type 3 secretion systems (T3SSs) are regulated by environmental signals and by feedback-control mechanisms (2–7). Feedback control means that secretory activity or assembly is directly linked to effector gene expression, maintaining the appropriate levels of effector gene expression (7, 8). Injectisome T3SSs are used by several pathogenic bacteria (e.g., Shigella, Pseudomonas, and Yersinia) to translocate protein effectors into the host cell and modulate virulence (2). In these systems, feedback control links the transcription of effector genes to secretory function (4, 5, 9). In bacteria with a single polar flagella (e.g., Campylobacter), feedback control links substrate gene expression to flagellar assembly (6, 7).
Mycobacteria are classified as diderm-mycolate bacteria (10–12). In addition to the cytoplasmic membrane (CM), mycobacteria have a mycolate-OM (MOM) with a lipid content that is distinct from other OMs (1, 10–12). The ESX/WSS secretion systems are a unique family of protein transporters that are thus far restricted to Gram-positive bacteria and mycobacteria (13–17). Although the ESX/WSS systems transport proteins across the CM, it is unclear how mycobacterial ESX proteins are translocated across the MOM (18).
ESX/WSS systems are functionally diverse with roles ranging from bacterial development to conjugation, metal homeostasis, and pathogenesis in different species (19–26). In mycobacteria, as many as five related ESX systems (ESX 1–5) can be encoded within the genome, with an additional system encoded on a plasmid for some species (27–29). Generally, ESX systems include several ESX conserved components (Eccs), small secreted Esx proteins, and several less conserved ESX-associated proteins (Esps) (13, 28, 30). Many of the Ecc proteins are localized to the CM, where they interact to form the ESX membrane complex (31–33).
The ESX-1 system is required for the virulence of mycobacterial pathogens, including Mycobacterium tuberculosis (M. tb), the causative agent of human tuberculosis, and the nontubercular species Mycobacterium marinum (26, 34–37). Expression of ESX-1 genes from M. tb in ESX-1–deficient M. marinum strains restores function and virulence, demonstrating that the two systems are functionally equivalent (37–39). In the host, pathogenic mycobacteria are taken up by professional phagocytes, including macrophages (40, 41). The ESX-1 system damages the phagosomal membrane, allowing bacterial interaction with the cytosol and survival. Mycobacteria lacking functional ESX-1 systems are attenuated and are retained in the phagosome (42–47). In the laboratory, the ESX-1 system promotes the secretion of several protein substrates into the culture media during growth (11, 35, 36, 39, 48–54).
Genes encoding ESX-1 substrates are directly regulated by several transcription factors in M. tb, including PhoP, EspR, and WhiB6 (53, 55–59). In M. marinum and in M. tb, WhiB6 is positively autoregulated and directly regulates the transcription of genes encoding several ESX-1 substrates and components (56, 58, 60). Recently, a model was proposed for M. marinum in which WhiB6 senses changes in the redox status of the environment and regulates the expression of ESX-1 genes. Thus, WhiB6 integrates a potential environmental signal and modulates ESX-1 secretion in M. marinum (60).
It is not known if ESX systems are regulated by feedback-control mechanisms akin to those understood for the T3SS in Gram-negative bacteria. We and others have observed that, when ESX systems in divergent mycobacterial species are inactivated by genetic disruption, substrate levels are reduced in the mycobacterial cell (35, 37, 52, 54, 61–67). We therefore hypothesized that ESX systems regulate the levels of ESX substrates within the cell by using a feedback-control mechanism. To test our hypothesis, we used transcriptional profiling, genetics, and quantitative proteomics in M. marinum to generate mechanistic evidence that the ESX-1 exporter is regulated by negative feedback control.
Results
ESX-1 Substrate Levels Are Regulated by EccCb1 in M. marinum.
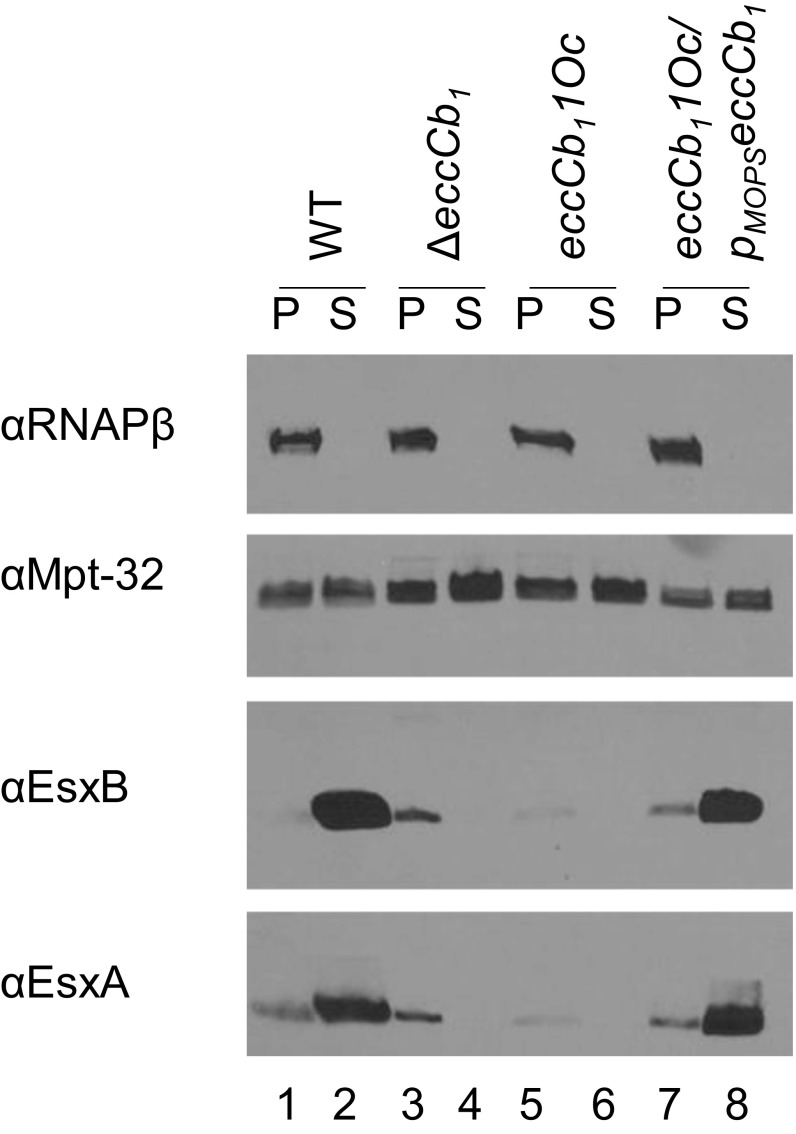
To define the mechanism of feedback control of the ESX-1 system in M. marinum, we recapitulated the observation that substrate levels are reduced in the absence of a functional ESX-1 system (35, 37, 52, 54, 61–67). We measured secretion of two major substrates, EsxA (ESAT-6) and EsxB (CFP-10), into the culture media during in vitro growth (Fig. 1). EsxA and EsxB were produced and secreted from the WT strain (Fig. 1, lanes 1 and 2). EccCb1 (Ecc Cb locus 1) is a component of the ESX-1-system (35, 36). M. tb and M. marinum strains lacking EccCb1 do not secrete ESX-1 substrates in vitro (35–37). The EsxA and EsxB proteins were produced in strains bearing a deletion in (Fig. 1, lane 3) or an ochre allele of eccCb1 [eccCb11(Oc) (68); Fig. 1, lane 5]. The eccCb11(Oc) allele encodes a premature stop codon following amino acid 48 of EccCb1, and does not produce EccCb1 protein (68). As expected, the EsxA and EsxB proteins were not secreted from the eccCb1 mutant strains (Fig. 1, lanes 4 and 6). Constitutive expression of eccCb1 from the synthetic mycobacterial optimal promoter (Mops) on an integrating plasmid (pMopseccCb1) restored the secretion of EsxA and EsxB from the eccCb11(Oc) strain (Fig. 1, lane 8). Importantly, the total amount of EsxA and EsxB protein produced and secreted from the WT strain (Fig. 1, lanes 1 and 2) appeared greater than the amount of EsxA and EsxB produced in the ESX-1-deficient strains (Fig. 1, lanes 3 and 4 or 5 and 6). From these data, we conclude that the total amount of the ESX-1 substrates, EsxA and EsxB, seems reduced in the absence of a functional eccCb1 gene.
Fig. 1.
ESX-1 substrates are regulated by feedback control. Western blot analysis of ESX-1 substrate production (“P,” pellet) and secretion (“S,” supernatant) during in vitro growth. RNAPβ is a control for mycobacterial lysis. MPT-32 is a Sec-secreted loading control. The experiment is representative of three independent biological replicates.
EccCb1 Regulates ESX-1 Associated Gene Expression in M. marinum.
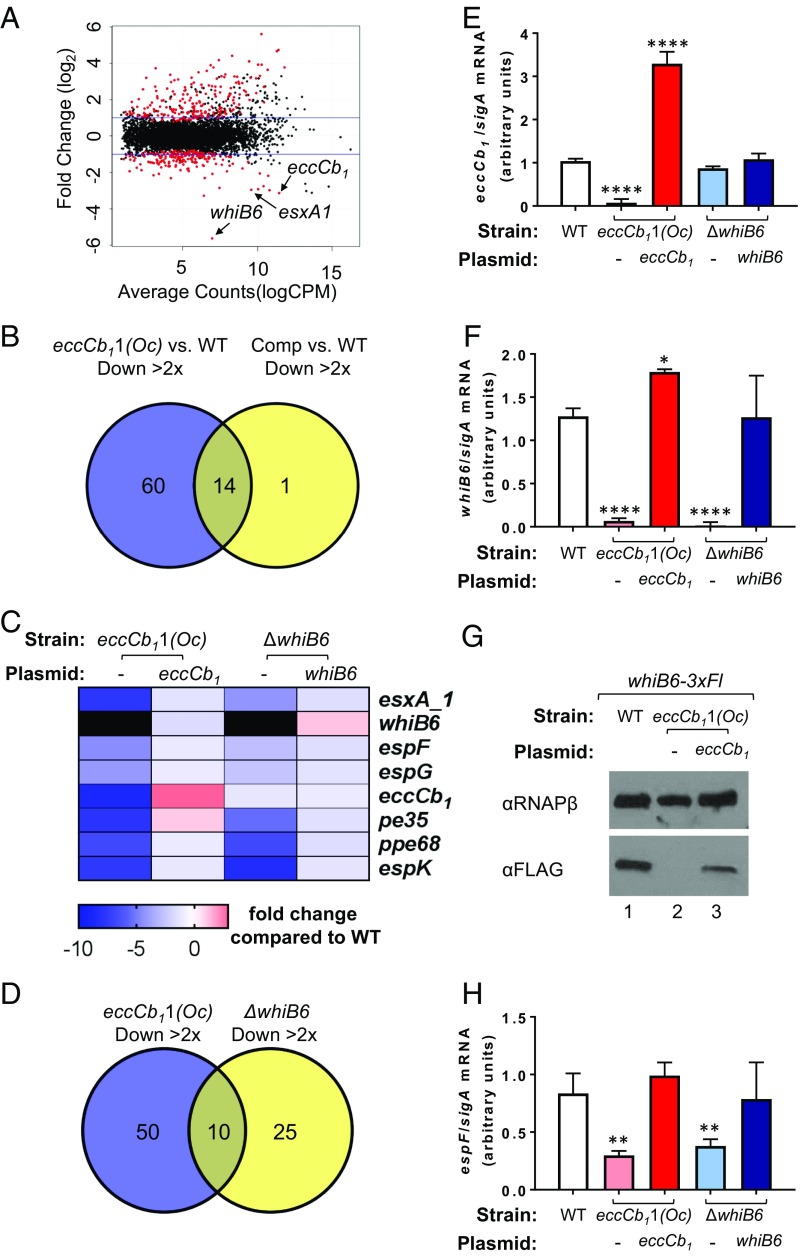
Feedback control directly links substrate gene expression to the secretory status or assembly of the protein transporter (5). We reasoned that the reduced levels of ESX-1 substrates observed in the absence of a functional eccCb1 gene could be the result of changes in substrate gene expression. To this end, transcriptional profiling analysis of the WT, eccCb11(Oc), and eccCb11(Oc) strain expressing a WT copy of the eccCb1 gene [eccCb11(Oc)/pMopseccCb1] was carried out to identify genes whose expression was dependent on the eccCb1 gene (Datasets S1 and S2). In the eccCb11(Oc) strain compared with the WT strain, 74 genes were down-regulated (more than twofold; q < 0.05, Fig. 2 A and B and Dataset S1B). Of these genes, 60 genes were not significantly down-regulated in the eccCb11(Oc)/pMopseccCb1 complemented strain relative to the WT strain, indicating that expression was restored by eccCb1 gene expression (Dataset S2). Therefore, these 60 genes represent genes putatively induced by eccCb1 gene expression (Dataset S3A). These genes include the MMAR_5437 gene, which encodes for the M. marinum ortholog of the whiB6 gene from M. tb (60, 69), several genes encoding substrates of the ESX-1 system, along with a copy of esxA_1 that is present elsewhere in the genome (selected ESX-associated genes are highlighted in Fig. 2C). Similarly, 92 genes were identified that were up-regulated (more than twofold; q < 0.05) in the eccCb11(Oc) strain compared with the WT, but that were not induced in the eccCb11(Oc)/pMopseccCb1 complemented strain (SI Appendix, Fig. S1, and Dataset S3B). These data demonstrate that gene expression is impacted, both positively and negatively, by the presence of the eccCb1 gene in M. marinum. Moreover, these findings support that EccCb1 is regulating (directly or indirectly) the expression of ESX-1 associated genes.
Fig. 2.
EccCb1 regulates whiB6 and ESX-1 substrate gene expression in M. marinum. (A) RNA-seq magnitude–amplitude plot of WT M. marinum vs. the eccCb11(Oc) strain. Selected genes that are down-regulated in the eccCb11(Oc) strain (whiB6, eccCb1, and esxA_1) are highlighted. Black dots indicate a lack of statistical significance, and red dots indicate statistical significance (q < 0.05). Complete data are presented in Dataset S1. (B) Venn diagram of genes down-regulated (more than twofold; q < 0.05) in the eccCb11(Oc) strain (Dataset S1C) or the complemented strain (Dataset S2C) relative to the WT strain. The putative EccCb1 regulated genes are shown in Dataset S3A. (C) Heat map of selected ESX-associated genes that are significantly down-regulated in the eccCb11(Oc) strain, several of which are also differentially regulated in ΔwhiB6 strain. Genes down-regulated greater than 10 fold are indicated in black. All plasmids in Fig. 2 include the pMops promoter. (D) Venn diagram of genes down-regulated (more than twofold; q < 0.05) in the eccCb11(Oc) strain but not the complemented strain (Dataset S3A), or in the whiB6 mutant strain but not the complemented strain (Dataset S3C), relative to the WT strain. The overlapping genes are enriched for ESX-associated genes (Dataset S3E). (E) Relative eccCb1 gene expression by qRT-PCR. The error bars represent propagated SD. The significance was defined by using an ordinary one-way ANOVA (P < 0.0001). eccCb1 levels were compared with those in the WT strain by using a Dunnett’s multiple comparison test (****P ≤ 0.0001). (F) whiB6 gene expression by qRT-PCR. Replicates and error bars are as in E. Significance was defined by using an ordinary one-way ANOVA (P < 0.0001). whiB6 levels were compared with the WT strain by using a Dunnett’s multiple comparison test (*P ≤ 0.05). (G) Western blot analysis of WhiB6-3xFL levels in the presence and absence of the eccCb1 gene from whole-cell lysates. RNAPβ was used as a loading control. The image is representative of three biological replicates. (H) espF gene transcription by qRT-PCR. Error bars represent SD. Significance was defined by using an ordinary one-way ANOVA (P = 0.0001). espF levels were compared with those in the WT strain by using a Dunnett’s multiple comparison test (**P ≤ 0.01). For E–G, all transcripts were normalized to the levels of sigA. Nonsignificant differences (P > 0.05) are not indicated. The qRT-PCR data represent the average of two biological replicates, each with two technical replicates.
The gene with the greatest transcriptional change in the eccCb11(Oc) strain was the whiB6 gene (MMAR_5437), which was down-regulated ∼50 fold compared with the WT strain. WhiB6 positively regulates the expression of several genes encoding ESX-1 substrates in M. marinum and M. tb (56, 59, 60). We generated a clean deletion of the whiB6 gene in M. marinum. We complemented the ΔwhiB6 strain by constitutively expressing the whiB6 gene from an integrating plasmid with the synthetic Mops promoter (ΔwhiB6/pMopswhiB6). Transcriptional profiling of the WT, ΔwhiB6, and ΔwhiB6/pMopswhiB6 complemented M. marinum strains was undertaken to identify genes with expression that is dependent on whiB6 (Datasets S4 and S5). In the ΔwhiB6 strain compared with the WT, 51 genes were down-regulated (more than twofold; q < 0.05; SI Appendix, Fig. S2 A and B, and Dataset S4C). Of these genes, 35 were not significantly down-regulated in the ΔwhiB6/pMopswhiB6 complemented strain relative to the WT strain, indicating that expression was restored by whiB6 gene expression (Dataset S3C). These genes represent putative WhiB6-activated genes. As expected, several genes whose expression was activated by WhiB6 encoded ESX-1 substrates (SI Appendix, Fig. S2C), which overlapped with those identified previously (60). These data support that WhiB6 induces the expression of ESX-1 associated genes. Several genes that were down-regulated in the ΔwhiB6 mutant strain appear to be involved in redox homeostasis and metal-ion physiology. A total of 34 genes were identified that were up-regulated (more than twofold; q < 0.05) in the ΔwhiB6 strain compared with the WT strain, but that were not induced in the complemented strain (SI Appendix, Fig. S2, and Dataset S3D), although these genes did not appear to be enriched for specific function or well-characterized pathways.
There was a significant overlap between the genes that were down-regulated in the eccCb11(Oc) and ΔwhiB6 strains, but not their respective complemented strains (Fig. 2D). These 10 genes include whiB6, esxA_1, and several genes encoding ESX-1 substrates or components (espF, espG, pe35, ppe68, and espK; a complete list of the overlapping genes is provided in Dataset S3E). Notably, eccCb1 gene expression was not down-regulated in the ΔwhiB6 strain. However, whiB6 gene expression was down-regulated in the eccCb11(Oc) strain. These data suggest that EccCb1, or a gene it regulates, is controlling whiB6 gene expression or activity and that WhiB6 is controlling expression of other coregulated genes including those encoding ESX-1 substrates.
We confirmed each of these findings by using quantitative RT-PCR (qRT-PCR). To this end, we isolated total RNA from the WT, eccCb11(Oc), eccCb11(Oc)/pMopseccCb1, ΔwhiB6, and ΔwhiB6/pMopswhiB6 strains. First, we confirmed that eccCb1 transcript levels were significantly reduced in the eccCb11(Oc) strain (P ≤ 0.0001; Fig. 2E). Constitutive expression of the eccCb1 gene in the eccCb11(Oc) strain restored eccCb1 transcript to levels significantly greater than those in the WT strain (P ≤ 0.0001). Expression of the eccCb1 gene was not significantly affected in the ΔwhiB6 or ΔwhiB6/pMopswhiB6 strains. From these data, we conclude that the eccCb1 gene is not regulated by WhiB6 in M. marinum.
We next tested if whiB6 gene expression was down-regulated in the eccCb11(Oc) strain. As shown in Fig. 2F, whiB6 transcript levels were significantly reduced in the eccCb11(Oc) strain [19-fold, comparable to the RNA sequencing (RNA-seq) data; P ≤ 0.0001] compared with the levels measured from the WT strain. Constitutive expression of the eccCb1 gene in the eccCb11(Oc) strain restored whiB6 transcript levels significantly greater than those detected in the WT strain (P ≤ 0.05). As a control for specificity, we measured whiB6 transcript levels in the ΔwhiB6 and ΔwhiB6/pMopswhiB6 strains. The levels of whiB6 transcript were significantly reduced in the ΔwhiB6 strain (P ≤ 0.0001) and restored to WT levels in the ΔwhiB6/pMopswhiB6 strain.
We sought to confirm that WhiB6 protein levels reflected the observed changes in whiB6 gene expression. Because we did not have an antibody for WhiB6, we integrated a whiB6 allele encoding for a WhiB6 protein with a C-terminal 3× FLAG epitope tag (whiB6-3xFl) into the ΔwhiB6 strain. The resulting strain encoded the whiB6-3xFl allele at the whiB6 locus (SI Appendix, Fig. S3). The WhiB6-3xFl protein binds DNA and activates transcription (59). We deleted the eccCb1 gene in the whiB6-3xFl strain and generated a complemented strain by introducing a plasmid constitutively expressing the eccCb1 gene. As shown in Fig. 2G, we detected WhiB6-3xFl in whole-cell lysates generated from the WT strain (Fig. 2G, lane 1). Consistent with the reduced levels of whiB6 transcription in strains lacking EccCb1, the WhiB6-3xFl protein was not detected in lysates from the ΔeccCb1 strain (Fig. 2G, lane 2). Expression of the eccCb1 gene from a plasmid restored the levels of WhiB6-3xFl protein (Fig. 2G, lane 3). These data indicate that WhiB6 protein levels are also reduced in the absence of EccCb1.
To confirm that ESX-1 substrate gene expression was reduced in the eccCb11(Oc) strain, we measured expression of the espF (MMAR_5440) and esxA substrate genes (11, 37, 51). As shown in Fig. 2H and SI Appendix, Fig. S4, we observed a significant decrease in espF (3.66-fold) and esxA (3.87-fold) transcript levels in the eccCb11(Oc) strain compared with the WT strain (P ≤ 0.01 for espF and P ≤ 0.0001 for esxA). Constitutive expression of the eccCb1 gene in the eccCb11(Oc) strain restored espF and esxA transcript levels to WT levels. The expression of the espF and esxA genes is positively regulated by WhiB6 in M. marinum (60). We measured a significant decrease in espF (2.99-fold) and esxA (4.57-fold) transcript levels in the ΔwhiB6 strain (P ≤ 0.01 for espF and P ≤ 0.0001 for esxA). Constitutive expression of the whiB6 gene in the ΔwhiB6 strain restored espF and esxA gene expression levels to WT levels. From these data, we conclude that, in the absence of EccCb1, espF and esxA gene expression is reduced, consistent with the RNA-seq data presented in Fig. 2 and the Western blot analysis data presented in Fig. 1. Together, these data demonstrate that EccCb1 regulates whiB6 and ESX-1 substrate gene expression in M. marinum.
Constitutive whiB6 Gene Expression Is Sufficient to Bypass EccCb1 Regulation of ESX-1 Genes.
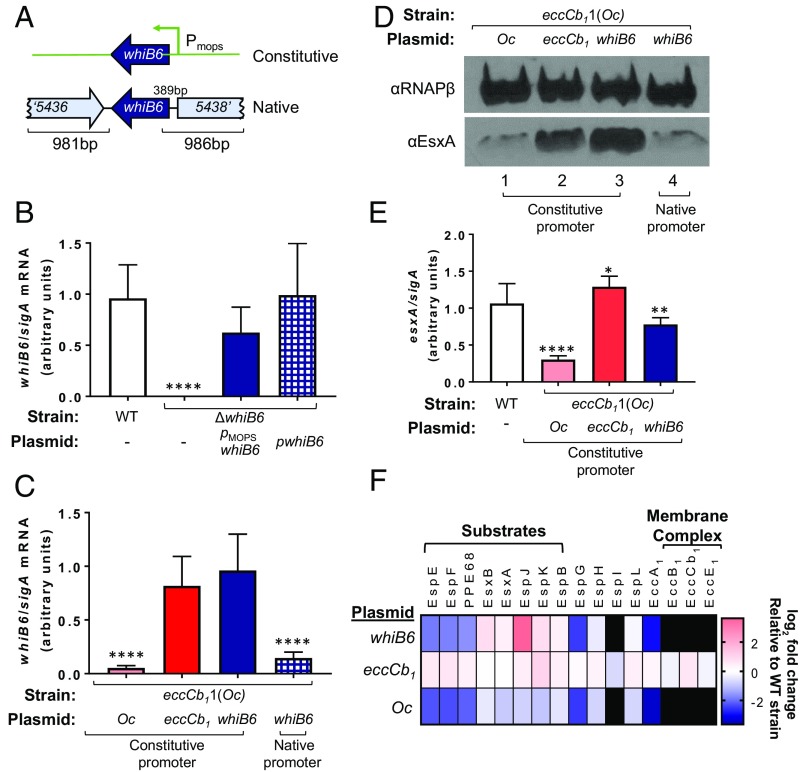
We sought to define how EccCb1 regulates whiB6 and ESX-1 gene expression. We hypothesized that EccCb1 regulated whiB6 gene expression at the level of transcription (Fig. 2), and that, in the absence of eccCb1, ESX-1 substrate gene expression was down-regulated by the loss of WhiB6. We reasoned that we could test this hypothesis by expressing whiB6 from a constitutive promoter (pMopswhiB6) or its native promoter (pwhiB6; Fig. 3A) in the absence of eccCb1. We hypothesized that, if the whiB6 promoter was the target of feedback control, we could bypass regulation by EccCb1 and restore ESX-1 substrates by expressing whiB6 from the constitutive promoter, but not the native promoter.
Fig. 3.
Constitutive expression of whiB6 is sufficient to bypass EccCb1-mediated regulation of ESX-1 genes. (A) Schematic of the constitutive (pMopswhiB6) and native (pwhiB6) whiB6 expression constructs. (B) whiB6 gene transcription by qRT-PCR. The whiB6 transcript was normalized to sigA. Data represent the average of three biological replicates, each performed in technical triplicate and normalized by using an interplate calibrator. Error bars represent the SD. Significance was defined by using an ordinary one-way ANOVA (P < 0.0001). whiB6 levels were compared with those in the WT strain by using a Šídák’s multiple comparison test (****P < 0.0001). Nonsignificant differences not indicated. “Oc” is the eccCb11(Oc) allele for Fig. 3. (C) whiB6 gene transcription measured as in B. Plasmid-borne genes expressed by the constitutive PMops promoter or the native promoter as indicated. Significance was defined by using an ordinary one-way ANOVA (P < 0.0001). whiB6 levels were compared across all strains by using a Tukey’s multiple comparison test. Significance compared with the eccCb11(Oc) strain with the pMopseccCb1 plasmid are shown. There were no significant differences between the eccCb11(Oc) strains bearing the pMopseccCb1 and the pMopswhiB6 plasmids (P = 0.5851) or between the eccCb11(Oc) strains bearing the pMopseccCb11(Oc) and pwhiB6 plasmids (P = 0.7704). (D) Western blot analysis of EsxA levels in whole-cell lysates generated from the eccCb11(Oc) strain. Plasmid-borne genes expressed by the constitutive PMops promoter (lanes 1–3) or the native promoter (lane 4) are indicated. RNAPβ was used as a loading control. The image shown is representative of three biological replicates. (E) esxA gene transcription by qRT-PCR. The esxA transcript was normalized to sigA. Data represent the average of four biological replicates, each with two technical replicates. Error bars represent the propagated SD. Significance was in comparison with WT as in C (**P ≤ 0.01, *P ≤ 0.05, and ****P ≤ 0.0001). In this panel, all genes were expressed from the constitutive promoter. (F) Quantification of ESX-1–associated proteins by using label-free proteomics. The strains are the same as E. All protein levels are represented as the log2 fold change compared with those measured in the WT strain. Black indicates that the proteins were quantified in the WT strain only. The experiment was performed on biological duplicates, each with two technical replicates.
The native expression plasmid was generated by introducing the whiB6 ORF and ∼1,000 bp upstream and downstream of the gene into an integrating plasmid (Fig. 3A). As shown in Fig. 3B, expression of whiB6 from the constitutive or native promoters restored the levels of whiB6 transcript to WT levels in the ΔwhiB6 strain. Similar to Fig. 2, whiB6 transcript levels were significantly reduced in the strain expressing the eccCb11(Oc) allele (P < 0.0001; Fig. 3C) compared with the levels measured from the strain expressing eccCb1. Constitutive, but not native, expression of the whiB6 gene restored whiB6 transcript levels in the eccCb11(Oc) strain. The levels of whiB6 expression in the strains with constitutive expression of the eccCb11(Oc) allele and native whiB6 gene expression were not significantly different. Together, these data demonstrate that the whiB6 promoter is likely the target of regulation by feedback control.
We compared the levels of EsxA protein in the eccCb11(Oc) strain expressing the eccCb11(Oc) allele [pMopseccCb11(Oc), vector control], the eccCb1 gene (pMopseccCb1, complemented), or the whiB6 gene from a constitutive promoter (pMopswhiB6) or its native promoter (pwhiB6). As shown in Fig. 3D, relative to the levels of EsxA protein in the eccCb11(Oc) strain (Fig. 3D, lane 1), expression of the eccCb1 gene increased the level of EsxA (Fig. 3D, lane 2). Constitutive expression of the whiB6 gene (Fig. 3D, lane 3) in the eccCb11(Oc) strain restored EsxA levels. Native expression of the whiB6 gene did not restore EsxA levels in the eccCb11(Oc) strain (Fig. 3D, lane 4). From these results, we conclude that constitutive expression of the whiB6 gene was sufficient to restore EsxA levels in M. marinum in the absence of EccCb1.
We further characterized the impact of constitutive expression of whiB6 in the eccCb11(Oc) strain. We hypothesized that restoration of whiB6 expression was increasing expression from the esxA gene. Consistent with this hypothesis, esxA gene expression was significantly reduced in the eccCb11(Oc) strain expressing the eccCb11(Oc) allele (P ≤ 0.0001) compared with the WT strain (Fig. 3E). Constitutive expression of the eccCb1 gene or the whiB6 gene increased esxA expression levels, even though both were still significantly different from the WT strain (P = 0.0432 and P = 0.0077, respectively). From these data, we conclude that expression of whiB6 from a constitutive promoter was sufficient to restore esxA gene expression in the absence of EccCb1.
WhiB6 regulates the expression of several genes encoding ESX-1 substrates in M. marinum (60) (Fig. 2). We sought to determine if constitutive expression of the whiB6 gene was sufficient to restore levels of known ESX-1 substrates in the eccCb11(Oc) strain. We performed global proteomics on whole-cell lysates of M. marinum WT, eccCb11(Oc), eccCb11(Oc)/pMopseccCb1, and eccCb11(Oc)/pMopswhiB6 (SI Appendix, Fig. S5, and Dataset S6). We identified 2,586 proteins at a 1% false discovery rate. Protein quantification was performed by using label-free-quantification (LFQ), which integrates the peak area of peptides corresponding to each protein (70).
We measured the levels of 20 ESX-1 proteins in the WT strain (Fig. 3F). Four proteins (MycP3, EccD1, PE35, and EccCa1) were confidently identified in only the WT strain, and the levels of these proteins were not measured in the eccCb11(Oc) strains. We identified 16 ESX-1 proteins (eight substrates and eight components) with reduced levels in the eccCb11(Oc) strain expressing the eccCb11(Oc) allele compared with the WT strain. We were surprised to find that, in addition to substrates encoded by WhiB6-regulated genes, several protein components of the ESX-1 system, whose gene expression is not regulated by WhiB6, were also reduced or were undetected in the eccCb11(Oc) strain compared with the WT strain. Interestingly, two proteins (EccB1 and EccE1) whose levels were reduced in the eccCb11(Oc) strain form the membrane complex with EccCb1 in the CM (56). All 16 proteins were restored to at least WT levels by expression of the eccCb1 gene in the eccCb11(Oc) strain. From these data, we conclude that, in the absence of EccCb1, the levels of ESX-1 substrates and components, including the membrane complex, are reduced.
Constitutive expression of whiB6 in the eccCb11(Oc) strain restored the levels of a subset of ESX-1 substrates (EsxA, EsxB, EspJ, EspK, and EspB) and components (EspH and EspL) to levels at or greater than the WT strain. From these data, we conclude that the loss of whiB6 gene expression, which results in a loss of WhiB6, causes the reduced levels of ESX-1 substrates in the eccCb11(Oc) strain (as observed in Fig. 1).
Interestingly, constitutive expression of whiB6 failed to restore the levels of three ESX-1 substrates (EspE, EspF, and PPE68), despite the fact that the espE, espF, and ppe68 gene expression is regulated by WhiB6 (60) (Fig. 2C). These data indicate that EspE, EspF, and PPE68 are also regulated independently of WhiB6 transcriptionally or posttranscriptionally.
As expected, constitutive expression of whiB6 also failed to restore the levels of EccCb1 in the eccCb11(Oc) strain. Interestingly, five additional components, two of which are in the ESX-1 membrane complex (EccB1, EccE1) were not restored by constitutive expression of the whiB6 gene. These data indicate that reduced levels of EspG, EspI, EccA1, EccB1, and EccE1 in the eccCb11(Oc) strain is independent of whiB6 gene expression.
Together, these data suggest that transcriptional regulation of whiB6 results in a reduction of ESX-1 substrates in the absence of EccCb1. Moreover, the loss of EccCb1 impacts the levels of components of the ESX-1 system, including the membrane complex, through an undescribed mechanism.
The ESX-1 Membrane Complex Regulates whiB6 Gene Expression in M. marinum.
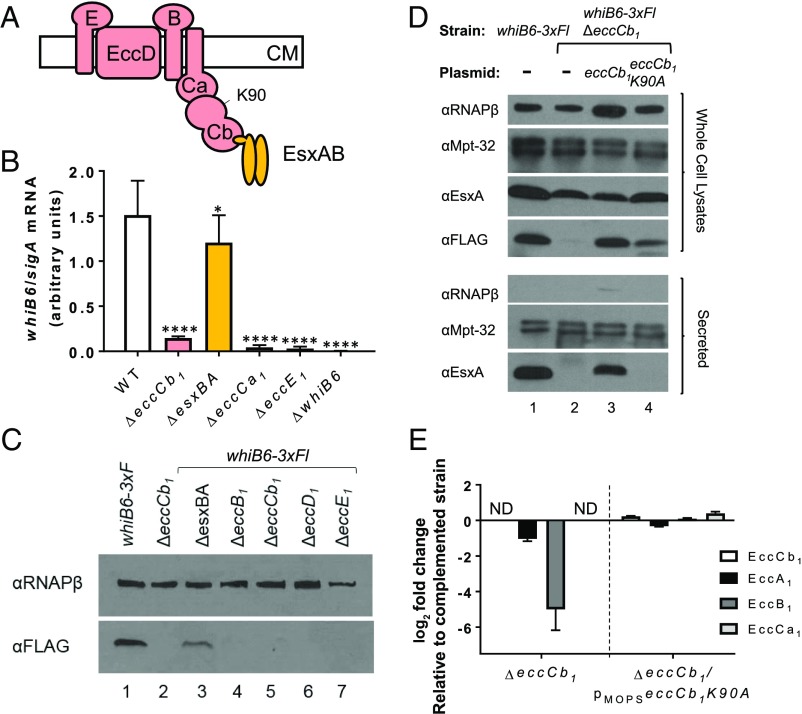
Based on the data in Fig. 3F, the loss of EccCb1 in the eccCb11(Oc) strain caused a reduction or loss of proteins that form the ESX-1 membrane complex (Fig. 4A) and other components. We hypothesized that regulation of whiB6 transcription was not specific to eccCb1. We tested if the deletion of other ESX-1 associated genes reduced whiB6 gene expression. Deletion of the esxBA genes, which encode for the EsxB and EsxA substrates, abrogates ESX-1 secretion (26, 35–37, 52). As shown in Fig. 4B, deletion of the esxBA genes significantly reduced whiB6 transcript levels compared with the WT strain (P < 0.05), but the levels of whiB6 gene transcription were significantly greater in the ΔesxBA strain compared with those measured in the ΔeccCb1 strain. Accordingly, we detected WhiB6-3xFl protein in the cell-associated fractions generated from the ΔesxBA strain (Fig. 4C). In contrast to the ΔesxBA strain, deletions in the eccCb1, eccCa1, eccE1, eccD1, or eccB1 genes (components of the membrane complex) significantly reduced whiB6 gene expression (Fig. 4B) or WhiB6-3xFl protein levels (Fig. 4C) compared with the WT strain, and similar to the eccCb11(Oc) strain (Fig. 2F). From these data, we conclude that loss of genes encoding ESX-1 components, but not the EsxBA substrates, promotes reduced whiB6 gene expression.
Fig. 4.
The membrane components regulate whiB6 gene expression in M. marinum. (A) Schematic representation of the ESX-1 membrane complex (pink) (31–33). The EsxA and EsxB substrates are shown in gold. The K90 residue is shown. (B) Relative whiB6 gene expression by qRT-PCR, normalized to sigA expression. Data shown represents three biological replicates, including seven technical replicates. Error bars represent propagated SD. Significance was defined by using an ordinary one-way ANOVA (P < 0.0001). whiB6 levels were compared with those in the WT strain by using a Dunnett’s multiple comparison test (*P ≤ 0.05 and ****P ≤ 0.0001). (C) Western blot analysis of WhiB6-3xFl levels in whole-cell lysates generated from strains lacking specific ESX-1–associated genes. Lanes 1 and 3–7 are strains with the whiB6-3xFl allele; lane 2 is WT for whiB6. RNAPβ is used as a loading control. eccCb1 genes are behind PMops. (D) Western blot analysis of whole-cell lysates (Upper) and secreted fractions (Lower) generated from the whiB6-3xFl ΔeccCb1 strains expressing the WT eccCb1 or eccCb1K90A alleles. RNAPβ serves as a lysis control. MPT-32 serves as a loading control. (E) Fold change in the levels of ESX-1 membrane proteins from LFQ proteomics. Three technical replicates and two biological duplicates were integrated and averaged. SE (percent coefficient of variation) was calculated for each technical triplicate. Propagation of error was performed to determine error for fold change. ND, not detected in the ΔeccCb1 strain. Protein ratios are reported as the log2 fold change compared with the complemented strain. Significance was determined as in Fig. 3.
Deletion of the esxBA genes abrogated ESX-1 secretion but did not phenocopy the changes in whiB6 transcription and protein levels measured in the ΔeccCb1 strain. Therefore, secretory status of the ESX-1 system was not likely to be the signal regulating whiB6 gene expression. Because whiB6 expression was not greatly affected in the absence of esxBA, EsxBA accumulation could be the signal for feedback regulation. To mimic EsxBA accumulation, we constitutively expressed esxBA genes encoding WT or EsxBA proteins that cannot be secreted [EsxBM98A (71)] and measured WhiB6-3xFl levels. Expression of the WT or mutant EsxBA proteins in the whiB6-3xFl strain did not appreciably reduce the levels of WhiB6-3xFl (SI Appendix, Fig. S6). Therefore, accumulation of EsxBA is likely not the signal for feedback regulation.
Another possibility consistent with our data was that the ESX-1 membrane complex regulates whiB6 gene expression. We sought a mutation in an ESX-1 component that abolished secretory activity, but did not affect the levels of the ESX-1 membrane proteins. The EccCb1 protein has two ATPase domains, referred to as ATPase domain 2 and 3 (72, 73). The K90 residue is required for the ATP binding by ATPase domain 2. Mutation of EccCb1K90 to A or T resulted in a stable but nonfunctional protein in M. tb (72, 73). We generated the M. marinum eccCb1K90A allele and constitutively expressed it in the whiB6-3xFlΔeccCb1 strain. As expected, the WT strain expressed the WhiB6-3xFl protein and secreted the EsxA substrate (Fig. 4D, lane 1). Deletion of eccCb1 resulted in the loss of WhiB6-3xFl protein and EsxA secretion (Fig. 4D, lane 2). WhiB6-3xFl levels and EsxA secretion were restored by expression of eccCb1 in the ΔeccCb1 strain (Fig. 4D, lane 3). Even though EsxA was not secreted from the ΔeccCb1 strain expressing EccCb1K90A allele, WhiB6-3xFl was detected (Fig. 4D, lane 4).
We next determined how the EccCb1K90A protein affected the levels of ESX-1 membrane proteins by using LFQ proteomics. As shown in Fig. 4E, we were unable to detect the EccCb1 protein in the ΔeccCb1 strain. The EccCb1 protein was identified at similar levels in the ΔeccCb1/pMopseccCb1K90A and the complemented strains (log2 fold change = 0.227). The levels of the ESX-1 conserved component proteins were reduced in the ΔeccCb1 strain compared with the complemented strain. EccA1 was reduced (log2 fold change = −1.037) in the ΔeccCb1 strain and slightly less so (log2 fold change = −0.321) in the ΔeccCb1/pMopseccCb1K90A strain compared with the complemented strain. The EccCa1 protein, which interacts directly with EccCb1 (35), was not detected in the ΔeccCb1 strain. The EccB1 protein was identified but at too low a level to quantify in the ΔeccCb1 strain. In contrast, both proteins were present in the ΔeccCb1/pMopseccCb1K90A strain and the complemented strain at similar levels (log2 fold changes = 0.393 and 0.099, respectively). We were unable to detect EccD1, and EccE1 was not reliably quantified in this analysis. These data indicate that expression of EccCb1K90A did not result in a significant reduction in the levels of the ESX-1 conserved components. Taken together, these data indicate that the presence or assembly of the ESX-1 membrane complex, not secretory activity, is the signal that regulates whiB6 gene expression.
Feedback Control Functions to Fine-Tune ESX-1 Gene Expression.
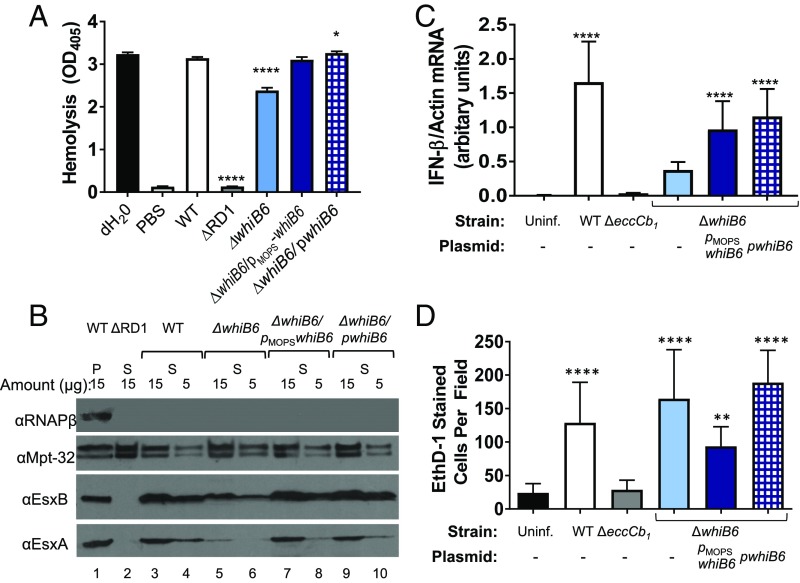
Genetic deletion does not reflect a physiological trigger for ESX-1 feedback control. Deletion of individual ESX-1 genes resulted in loss of whiB6 gene expression (Fig. 4 B and C). However, in strains bearing mutation or deletion of eccCb1 or deletion of whiB6, substrate gene expression was reduced, not abrogated. We therefore reasoned that characterizing ESX-1 secretion levels from the ΔwhiB6 strain could inform our understanding of feedback control. It was previously reported by Chen et al. (60) that replacement of the whiB6 gene with the hygromycin resistance gene (whiB6::hyg) in M. marinum resulted in a loss of ESX-1 secretion. However, Chen et al. observed that whiB6::hyg M. marinum retained hemolytic activity and virulence, both of which require a functional ESX-1 system. We hypothesized that feedback control functions to fine-tune the levels of ESX-1 secretion, rather than shutting the system off.
Consistent with the report by Chen et al. (60), the hemolytic activity of the ΔwhiB6 strain was slightly, but significantly (P ≤ 0.0001), reduced compared with the WT strain (Fig. 5A). Expression of the whiB6 gene from the constitutive or native promoters restored hemolytic activity to WT levels. In contrast, the ΔRD1 strain, which bears a deletion of several ESX-1 genes and is ESX-1–deficient, had hemolytic activity similar to the PBS (i.e., no bacteria) control. These data indicate that the ESX-1 system in the ΔwhiB6 strain is functional.
Fig. 5.
Feedback control fine-tunes ESX-1 function. (A) Sheep RBC (sRBC) hemolysis assay performed on three biological replicates, each in technical triplicate. The data represent the average between the three biological replicates. Error bars represent the propagated SD. Significance was determined by using an ordinary one-way ANOVA (P < 0.0001). The levels of hemolysis were compared with those of the WT strain by using a Dunnett’s multiple comparison test (****P ≤ 0.0001 and *P ≤ 0.05). Nonsignificant changes (P > 0.05) are not indicated. (B) Western blot analysis of EsxA and EsxB secretion. Lane 1 is whole-cell lysates from the WT strain. Lanes 2–10 are supernatant fractions. The amount loaded in each lane is indicated in micrograms. RNAPβ is a lysis control. MPT-32 serves as a loading control. (C) qRT-PCR measuring the induction of the type I IFN response by RAW 264.7 cells 4 h after infection with M. marinum at an MOI of 5. IFN-β expression was normalized to actin expression. Data are representative of six biological replicates, each with two technical replicates. Significance was determine by using an ordinary one-way ANOVA (P < 0.0001) followed by a Tukey’s multiple comparison test. Significance is relative to the uninfected control. The levels of IFN-β induction were not significantly different from the ΔeccCb1 and ΔwhiB6 strains (P > 0.9999 for ΔeccCb1 and P = 0.1021 for ΔwhiB6). (D) Quantification of cytolysis of RAW 264.7 cells. Five random fields were counted from each of three wells. The 15 resulting counts were averaged. The error bar represents SD between these counts. The experiment shown is representative of three biological replicates. Significance was determined by using an ordinary one-way ANOVA (P < 0.0001) followed by a Tukey’s multiple comparison test. Significance shown is based on the comparison against the uninfected strain (**P = 0.001). Nonsignificant changes are not shown. The number of EthD-1 cells was not significantly different between the WT and the ΔwhiB6 strains.
We next measured secretion in the absence of the whiB6 gene. Clean deletion of the whiB6 gene did not abrogate ESX-1 substrate secretion in vitro (SI Appendix, Fig. S7). However, with 20 µg of secreted protein loaded for analysis, the levels of EsxA secretion appeared reduced in the ΔwhiB6 strain compared with the WT and whiB6/pMopswhiB6 complemented strains. We verified the secretion of EsxA and EsxB by loading a series of concentrations of the secreted fraction (15 µg and 5 µg; Fig. 5B). Deletion of the whiB6 gene resulted in a reduction, but not a loss of EsxA and EsxB secretion (Fig. 5B, lanes 5 and 6). The levels of EsxA and EsxB in the secreted fraction were restored by whiB6 expression from the constitutive or native promoters (Fig. 5B, lanes 7–10). We conclude that deletion of the whiB6 gene does not abrogate ESX-1 function. Moreover, constitutive or native expression of whiB6 functionally complements the ΔwhiB6 strain.
To determine the physiological relevance of the feedback control, we tested how reduced ESX-1 function affected virulence of M. marinum in a macrophage infection model. In the macrophage, the ESX-1 system promotes cytosolic access, which leads to induction of the type I IFN, IFN-β, and lysis of the macrophage (42, 46, 47, 74). We infected RAW 264.7 cells with M. marinum at a multiplicity of infection (MOI) of 5 and measured induction of IFN-β 4 h after infection by using qRT-PCR. As shown in Fig. 5C, the WT M. marinum strain significantly induced expression of IFN-β compared with the uninfected control (P = 0.0001). Induction of IFN-β by the ΔeccCb1 and ΔwhiB6 strains was not significantly different from the uninfected control. Expression of whiB6 from the constitutive or the native promoter in the ΔwhiB6 strain restored induction of IFN-β relative to the uninfected control (P < 0.0001). The levels of induction between the WT and the ΔwhiB6/pMopswhiB6 strain, but not the ΔwhiB6/pwhiB6 strains, were significantly different from each other (P = 0.0166 and P = 0.1425, respectively). These data indicate that the ΔwhiB6 strain exhibits attenuated induction of IFN-β transcript compared with the WT strain.
We next measured macrophage cytolysis 24 h after infection (SI Appendix, Fig. S8). Ethidium homodimer (EthD-1) is a nucleic acid stain that is taken up only by cells with damaged cell membranes. In permeabilized cells, EthD-1 binds DNA and emits a red fluorescent signal. We counted the number of red cells per field to measure the cytolytic activity of each M. marinum strain (Fig. 5D; images in SI Appendix, Fig. S8). Infection of the RAW 264.7 cells with the WT, ΔwhiB6,ΔwhiB6/pMopswhiB6, and ΔwhiB6/pwhiB6 strains caused significant increases in cytolysis (red cells, P < 0.0001 for WT, ΔwhiB6, and ΔwhiB6/pwhiB6 strains, and P = 0.0010 for ΔwhiB6/pMopswhiB6) compared with the ΔeccCb1 strain and the uninfected control. The cytolytic activities of the WT and ΔwhiB6 strains were not significantly different from each other (P = 0.2706). Together, these data indicate that the ΔwhiB6 strain was attenuated for ESX-1 function during early stages, but not during later stages, of infection in the macrophage model. We interpret these data to mean that feedback control, which results in a loss of WhiB6, functions to fine-tune ESX-1 secretion in M. marinum.
Discussion
Our study demonstrates that the ESX-1 system in M. marinum is regulated by negative feedback control, linking the expression of the whiB6 gene and genes encoding ESX-1 substrates to the status of the ESX-1 membrane complex. Collectively, our experimental data support a model for feedback regulation of ESX-1 in M. marinum that fine-tunes the levels of ESX-1 substrates. When the levels of ESX-1 components, most likely the membrane complex, are reduced, whiB6 gene expression is down-regulated. Because expression of whiB6 from a constitutive promoter, but not from the native promoter, restored whiB6 expression and ESX-1 substrate levels in the absence of eccCb1, the whiB6 promoter is likely the target of feedback control. Reduced whiB6 gene expression, which leads to a loss of WhiB6 protein, results in the down-regulation of genes encoding ESX-1 substrates and a reduction of the substrate pool within the cell.
Our model agrees with published evidence from other groups. Several groups have demonstrated that substrate levels are reduced or do not accumulate in various strains lacking a functional ESX-1 exporter in Mycobacterium smegmatis (61), a distantly related nonpathogenic species, or in M. tb (35, 62) and M. marinum (37, 52, 54). Conversely, substrate accumulation has also been reported in M. marinum (51, 52), M. tb (50), and M. smegmatis (75). In all three species, there is a whiB6 ortholog (MSMEG_0051, Rv3862c) encoded upstream of the ESX-1 system (56). Lack of substrate accumulation has also been observed in strains deficient for ESX-3 (63) or ESX-5 export (64–66, 76). Genes encoding the ESX-5 system are also regulated by transcription factors (21, 77), which sense and respond to the environment, and may be targets of feedback control. Together, these observations suggest that ESX systems in different mycobacterial species are regulated by feedback control, although further study is required.
Our study also indicates that feedback control is not regulated by transcription alone. Although espF gene expression is regulated by WhiB6, expression of whiB6 in the absence of eccCb1 did not restore EspF protein (Fig. 3). Therefore, it is likely that posttranscriptional regulation is also contributing to feedback control. Indeed, the idea of posttranscriptional regulation of the levels of ESX substrates and secretion itself has been raised before. For example, the levels of the ESX-5 substrate LipY appear to be posttranscriptionally regulated (64). Moreover, the MycP1 protease regulates ESX-1 secretion posttranscriptionally. Loss of protease activity does not disrupt ESX-1 function, but results in increased levels of secreted ESX-1 substrates. Thus, the protease activity of MycP1 negatively regulates the secretion of, but not the levels of, the ESX-1 substrates within the cell (32, 78).
In the absence of the eccCb1 gene, we did not detect the components of the ESX-1 system that reside in the membrane (EccCa1, EccE1). It was previously demonstrated that five Ecc proteins (EccE, EccD, EccB, EccCa, and EccCb), from the ESX-1 and ESX-5 systems, form a complex in the CM (32, 33). The structure of the ESX-5 membrane complex from Mycobacterium xenopi was recently solved (31). Our data indicate that, in M. marinum, the loss of a single component of this complex (EccCb1) promotes the loss or reduction of several membrane components, in line with our previous study (52). These findings agree with reports that the deletion of other Eccs (mycosins or EccE) lead to reduced levels of the other components in the CM (31–33).
Our findings are inconsistent with a subset of those reported by Chen et al. (60). In that report, the authors demonstrated that disruption of the whiB6 gene in M. marinum M strain abolished the secretion of ESX-1 substrates in vitro (60). The data in Fig. 5B contradict this conclusion. We observed that clean deletion of the whiB6 gene resulted in a reduction, but not a loss, of ESX-1 activity. Indeed, we observed secretion of EsxA and EsxB and hemolysis in the ΔwhiB6 strain to levels significantly higher than in ESX-1–deficient strains. It is likely that in the study of Chen et al. (60), too little protein was analyzed to detect protein secretion by Western blot analysis. Secretion at lower than the levels of detection by Western blot analysis are sufficient to promote hemolysis and virulence (63, 79–81). The H37Rv M. tb strain is virulent despite reduced levels of ESX-1 secretion (56). Likewise, the ΔwhiB6 strain retained virulence in the macrophage model of infection. The reduced levels of ESX-1 secretion in the ΔwhiB6 strain may have resulted in delayed ESX-1–associated virulence in the macrophage model. Chen et al. reported that the whiB6::hyg M. marinum strain retained virulence in a zebrafish model of infection (60). Our study indicates that feedback control functions as a dial to fine-tune ESX-1 protein secretion, rather than as a switch.
Our findings are reminiscent of established mechanisms of feedback regulation used in controlling T3SS in Gram-negative pathogens (reviewed in refs. 2, 4, 5). We found that, like injectisome systems, posttranscriptional mechanisms are also involved (82). Our model is also similar to feedback regulation of the flagellar T3SS system in Campylobacter jejuni. In C. jejuni, the assembly of the T3SS and the surrounding apparatus in the CM regulates substrate gene expression (6, 7, 83). Flagellar proteins localized beyond the CM are substrates of the T3SS (9). Importantly, mutations that abolish T3 secretory activity but allow the assembly of the apparatus promote substrate gene expression (7). Because the EccCb1K90A protein abolished ESX-1 secretion, but did not appreciably reduce the levels of WhiB6 or the other ESX-1 membrane proteins, the presence or assembly of the ESX-1 membrane complex likely regulates whiB6 gene expression. In T3SS, feedback control fine-tunes effector and substrate pools (8). Feedback control promotes the maintenance of the effector pool, preventing depletion or accumulation of effectors within the cell that could impact bacterial survival (8). Our findings also indicate that feedback control is a widespread regulatory mechanism of bacterial protein secretion, spanning at least the evolutionarily divergent T3SS and ESX-1 systems.
More work is needed to further elucidate this signaling pathway. It is unclear what physiological conditions would lead to a reduction in ESX-1 components. The ESX-1 membrane complex could represent a regulatory checkpoint in the assembly of the ESX-1 exporter (7). When the checkpoint is successfully reached, whiB6 and ESX-1 substrate gene expression are increased. The ESX-1 substrates may contribute to assembly of the functional ESX-1 apparatus (73). If the membrane complex forms incorrectly, the levels of the ESX-1 membrane proteins could be reduced, leading to a loss of whiB6 gene expression and reduced substrate gene expression. Alternatively, the loss of the complex may be a response to an upstream signal. It is unknown how the loss of the membrane complex signals to reduce whiB6 transcription. The EccCb1 protein is not likely regulating whiB6 gene expression directly. Because WhiB6 is positively autoregulated (60), WhiB6 may sense the loss of the ESX-1 membrane complex directly. WhiB6 is one of seven WhiB-like (Wbl) transcription factors in Mycobacterium (84, 85). Wbl transcription factors sense and respond to diatomic gasses and redox signals (e.g., NO and O2) (86). WhiB6 senses changes in redox homeostasis in M. marinum (60). However, we do not think WhiB6 senses the ESX-1 membrane complex directly. Constitutive whiB6 expression bypassed feedback regulation of ESX-1 substrates (Fig. 3). These data strongly suggest that the WhiB6 protein is functional in the absence of ESX-1 (60, 86). The assembly of the T3S flagellar apparatus regulates substrate gene expression through the FlgSR two-component system (7, 83, 87). There is likely a regulatory pathway that connects the ESX-1 membrane complex to whiB6 gene expression. We envision a negative regulator that represses whiB6 transcription or a positive regulator that no longer activates whiB6 transcription in the absence of eccCb1 and the membrane complex. Indeed, whiB6 is directly regulated at the level of transcription in M. tb by the PhoPR two component system, by WhiB3, and by the orphan response regulator Rv0818 (56). These regulators may be candidates for the signal transduction pathway connecting the ESX-1 membrane complex to the whiB6 promoter in M. marinum.
In conclusion, we have identified a signaling pathway in M. marinum that regulates the levels of ESX-1 substrates. To our knowledge, this is the first report of a regulatory pathway that links the Ecc proteins to expression of ESX substrate genes at the ESX-1 locus. Our study explains, at least in part, the fundamental mechanism underlying how ESX-1 substrate pools are fine-tuned and why substrates do not accumulate in the absence of a functional ESX-1 exporter.
Materials and Methods
All M. marinum strains used in this study were generated from the M strain (American Type Culture Collection BAA-535) using the allelic exchange as described previously (68). All strains and plasmids are described in SI Appendix, Table S1, and all oligonucleotide primers are listed in SI Appendix, Table S2. ESX-1 secretion assays were performed as previously described (88). Mycobacterial whole-cell lysates were prepared for bottom-up proteomics and analyzed by LC/MS/MS as in previous publications (44, 45). Database identification and LFQ were performed as described previously (70). RNA was purified by using the RNeasy Mini Kit, and quality was assessed by using an Agilent Bioanalyzer 2100 and the Agilent RNA 6000 Nano Reagent kit (16S rRNA chip) at the University of Notre Dame Genomics Facility. RNA-seq was performed as described previously (89, 90), and analyzed by using the SPARTA software package (91). Genes with an average log2 cpm < 5 were filtered from the final differential gene expression lists (Datasets S1–S5). The transcriptional profiling data are available at the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE99632). Details regarding cDNA synthesis and qRT-PCR analysis are reported in SI Appendix, Tables S2 and S3. Hemolysis assays were performed as described previously (88), except bacterial cells were incubated with sRBCs for 1.5 h at 30 °C. Readings were performed on a SpectraMax M5 plate reader (Molecular Devices) and analyzed by using SoftMax Pro-5 software (Molecular Devices). Biological and technical replicates are indicated in the figure legends. Macrophage infections and cytotoxicity assays were performed exactly as described previously (88). Counts were performed exactly as described previously (69) by using ImageJ. The cytokine assays were performed exactly as described previously (69, 88), with details reported in SI Appendix, Tables S2 and S3. Statistical analyses are described in the appropriate figure legends. A full explanation of study methods is provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Genomics and Bioinformatics Facility and the Mass Spectrometry and Proteomics Facility at the University of Notre Dame for their support of this research, Dr. Benjamin Johnson for bioinformatics support, and Dr. David Hendrixson for helpful discussion. The research reported in this study was supported by National Institute of Allergies and Infectious Diseases/National Institutes of Health (NIH) Awards R01AI106872 (to P.A.C.) and R01AI116605 (to R.B.A.) and a Graduate Student Fellowship from the Eck Institute for Global Health at the University of Notre Dame (to M.J.F.). This study is based on work supported by a National Science Foundation Graduate Research Fellowship under Grant DGE-1313583 (to R.E.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE99632).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710167114/-/DCSupplemental.
References
- 1.Chagnot C, Zorgani MA, Astruc T, Desvaux M. Proteinaceous determinants of surface colonization in bacteria: Bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol. 2013;4:303. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng W, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 3.McCaw ML, Lykken GL, Singh PK, Yahr TL. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol. 2002;46:1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 4.Diaz MR, King JM, Yahr TL. Intrinsic and extrinsic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Front Microbiol. 2011;2:89. doi: 10.3389/fmicb.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brutinel ED, Yahr TL. Control of gene expression by type III secretory activity. Curr Opin Microbiol. 2008;11:128–133. doi: 10.1016/j.mib.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boll JM, Hendrixson DR. A regulatory checkpoint during flagellar biogenesis in Campylobacter jejuni initiates signal transduction to activate transcription of flagellar genes. MBio. 2013;4:e00432-13. doi: 10.1128/mBio.00432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller VL. Connections between transcriptional regulation and type III secretion? Curr Opin Microbiol. 2002;5:211–215. doi: 10.1016/s1369-5274(02)00303-x. [DOI] [PubMed] [Google Scholar]
- 9.Büttner D. Protein export according to schedule: Architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sani M, et al. Direct visualization by cryo-EM of the mycobacterial capsular layer: A labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bansal-Mutalik R, Nikaido H. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc Natl Acad Sci USA. 2014;111:4958–4963. doi: 10.1073/pnas.1403078111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdallah AM, et al. Type VII secretion–Mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 14.Houben EN, Korotkov KV, Bitter W. Take five–type VII secretion systems of mycobacteria. Biochim Biophys Acta. 2014;1843:1707–1716. doi: 10.1016/j.bbamcr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Unnikrishnan M, Constantinidou C, Palmer T, Pallen MJ. The enigmatic Esx proteins: Looking beyond mycobacteria. Trends Microbiol. 2017;25:192–204. doi: 10.1016/j.tim.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Pallen MJ, Chaudhuri RR, Henderson IR. Genomic analysis of secretion systems. Curr Opin Microbiol. 2003;6:519–527. doi: 10.1016/j.mib.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Pallen MJ. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 18.Bosserman RE, Champion PA. Esx systems and the mycobacterial cell envelope: What’s theconnection? J Bacteriol. 2017;199:e00131-17. doi: 10.1128/JB.00131-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegrist MS, et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci USA. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fyans JK, Bignell D, Loria R, Toth I, Palmer T. The ESX/type VII secretion system modulates development, but not virulence, of the plant pathogen Streptomyces scabies. Mol Plant Pathol. 2013;14:119–130. doi: 10.1111/j.1364-3703.2012.00835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott SR, Tischler AD. Phosphate responsive regulation provides insights for ESX-5 function in Mycobacterium tuberculosis. Curr Genet. 2016;62:759–763. doi: 10.1007/s00294-016-0604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coros A, Callahan B, Battaglioli E, Derbyshire KM. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol. 2008;69:794–808. doi: 10.1111/j.1365-2958.2008.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottai D, et al. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol. 2012;83:1195–1209. doi: 10.1111/j.1365-2958.2012.08001.x. [DOI] [PubMed] [Google Scholar]
- 24.Korea CG, et al. Staphylococcal Esx proteins modulate apoptosis and release of intracellular Staphylococcus aureus during infection in epithelial cells. Infect Immun. 2014;82:4144–4153. doi: 10.1128/IAI.01576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kneuper H, et al. Heterogeneity in ess transcriptional organization and variable contribution of the Ess/type VII protein secretion system to virulence across closely related Staphylocccus aureus strains. Mol Microbiol. 2014;93:928–943. doi: 10.1111/mmi.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu T, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci USA. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton-Foot M, Warren RM, Sampson SL, van Helden PD, Gey van Pittius NC. The plasmid-mediated evolution of the mycobacterial ESX (type VII) secretion systems. BMC Evol Biol. 2016;16:62. doi: 10.1186/s12862-016-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2:RESEARCH0044. doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ummels R, et al. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. MBio. 2014;5:e01744-14. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bitter W, et al. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009;5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beckham KS, et al. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat Microbiol. 2017;2:17047. doi: 10.1038/nmicrobiol.2017.47. [DOI] [PubMed] [Google Scholar]
- 32.van Winden VJ, et al. Mycosins are required for the stabilization of the ESX-1 and ESX-5 type VII secretion membrane complexes. MBio. 2016;7:e01471-16. doi: 10.1128/mBio.01471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houben EN, et al. Composition of the type VII secretion system membrane complex. Mol Microbiol. 2012;86:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 34.Volkman HE, et al. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci USA. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guinn KM, et al. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao LY, et al. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- 38.Conrad WH, et al. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc Natl Acad Sci USA. 2017;114:1371–1376. doi: 10.1073/pnas.1620133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaughlin B, et al. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 2007;3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suter E. The multiplication of tubercle bacilli within normal phagocytes in tissue culture. J Exp Med. 1952;96:137–150. doi: 10.1084/jem.96.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner WH. Host-parasite interactions with peritoneal macrophages of mice and rats in vitro and in vivo. Infect Immun. 1975;12:1295–1306. doi: 10.1128/iai.12.6.1295-1306.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simeone R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamm LM, et al. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med. 2003;198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Wel N, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 45.Houben D, et al. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 2012;14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 46.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacGurn JA, Raghavan S, Stanley SA, Cox JS. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol Microbiol. 2005;57:1653–1663. doi: 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- 49.Berthet FX, Rasmussen PB, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10) Microbiology. 1998;144:3195–3203. doi: 10.1099/00221287-144-11-3195. [DOI] [PubMed] [Google Scholar]
- 50.Fortune SM, et al. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci USA. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Champion PA, Champion MM, Manzanillo P, Cox JS. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol Microbiol. 2009;73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Champion MM, Williams EA, Pinapati RS, Champion PA. Correlation of phenotypic profiles using targeted proteomics identifies mycobacterial esx-1 substrates. J Proteome Res. 2014;13:5151–5164. doi: 10.1021/pr500484w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature. 2008;454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlsson F, Joshi SA, Rangell L, Brown EJ. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog. 2009;5:e1000285. doi: 10.1371/journal.ppat.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anil Kumar V, et al. EspR-dependent ESAT-6 protein secretion of Mycobacterium tuberculosis requires the presence of virulence regulator PhoP. J Biol Chem. 2016;291:19018–19030. doi: 10.1074/jbc.M116.746289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solans L, et al. A specific polymorphism in Mycobacterium tuberculosis H37Rv causes differential ESAT-6 expression and identifies WhiB6 as a novel ESX-1 component. Infect Immun. 2014;82:3446–3456. doi: 10.1128/IAI.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broset E, Martín C, Gonzalo-Asensio J. Evolutionary landscape of the Mycobacterium tuberculosis complex from the viewpoint of PhoPR: Implications for virulence regulation and application to vaccine development. MBio. 2015;6:e01289-15. doi: 10.1128/mBio.01289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rustad TR, et al. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 2014;15:502. doi: 10.1186/s13059-014-0502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minch KJ, et al. The DNA-binding network of Mycobacterium tuberculosis. Nat Commun. 2015;6:5829. doi: 10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Z, et al. Mycobacterial WhiB6 differentially regulates ESX-1 and the dos regulon to modulate granuloma formation and virulence in zebrafish. Cell Rep. 2016;16:2512–2524. doi: 10.1016/j.celrep.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 61.Converse SE, Cox JS. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol. 2005;187:1238–1245. doi: 10.1128/JB.187.4.1238-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brodin P, et al. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun. 2006;74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegrist MS, et al. Mycobacterial Esx-3 requires multiple components for iron acquisition. MBio. 2014;5:e01073-14. doi: 10.1128/mBio.01073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daleke MH, et al. Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. J Biol Chem. 2011;286:19024–19034. doi: 10.1074/jbc.M110.204966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abdallah AM, et al. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 2009;73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- 66.Abdallah AM, et al. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 2006;62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- 67.Daleke MH, et al. Specific chaperones for the type VII protein secretion pathway. J Biol Chem. 2012;287:31939–31947. doi: 10.1074/jbc.M112.397596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams EA, et al. A nonsense mutation in Mycobacterium marinumthat is suppressible by a novel mechanism. Infect Immun. 2017;85:e00653-16. doi: 10.1128/IAI.00653-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kapopoulou A, Lew JM, Cole ST. The MycoBrowser portal: A comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 2011;91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 70.Braselmann E, Chaney JL, Champion MM, Clark PL. DegP chaperone suppresses toxic inner membrane translocation intermediates. PLoS One. 2016;11:e0162922. doi: 10.1371/journal.pone.0162922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Champion PA, Stanley SA, Champion MM, Brown EJ, Cox JS. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science. 2006;313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- 72.Ramsdell TL, Huppert LA, Sysoeva TA, Fortune SM, Burton BM. Linked domain architectures allow for specialization of function in the FtsK/SpoIIIE ATPases of ESX secretion systems. J Mol Biol. 2015;427:1119–1132. doi: 10.1016/j.jmb.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenberg OS, et al. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell. 2015;161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 75.Wirth SE, et al. Polar assembly and scaffolding proteins of the virulence-associated ESX-1 secretory apparatus in mycobacteria. Mol Microbiol. 2012;83:654–664. doi: 10.1111/j.1365-2958.2011.07958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daleke MH, et al. General secretion signal for the mycobacterial type VII secretion pathway. Proc Natl Acad Sci USA. 2012;109:11342–11347. doi: 10.1073/pnas.1119453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elliott SR, Tischler AD. Phosphate starvation: A novel signal that triggers ESX-5 secretion in Mycobacterium tuberculosis. Mol Microbiol. 2016;100:510–526. doi: 10.1111/mmi.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohol YM, et al. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen JM, et al. Phenotypic profiling of Mycobacterium tuberculosis EspA point mutants reveals that blockage of ESAT-6 and CFP-10 secretion in vitro does not always correlate with attenuation of virulence. J Bacteriol. 2013;195:5421–5430. doi: 10.1128/JB.00967-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Champion PA. Disconnecting in vitro ESX-1 secretion from mycobacterial virulence. J Bacteriol. 2013;195:5418–5420. doi: 10.1128/JB.01145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kennedy GM, Hooley GC, Champion MM, Mba Medie F, Champion PA. A novel ESX-1 locus reveals that surface-associated ESX-1 substrates mediate virulence in Mycobacterium marinum. J Bacteriol. 2014;196:1877–1888. doi: 10.1128/JB.01502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schulmeyer KH, Yahr TL. Post-transcriptional regulation of type III secretion in plant and animal pathogens. Curr Opin Microbiol. 2017;36:30–36. doi: 10.1016/j.mib.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Joslin SN, Hendrixson DR. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J Bacteriol. 2009;191:2656–2667. doi: 10.1128/JB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Larsson C, et al. Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One. 2012;7:e37516. doi: 10.1371/journal.pone.0037516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alam MS, Garg SK, Agrawal P. Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. FEBS J. 2009;276:76–93. doi: 10.1111/j.1742-4658.2008.06755.x. [DOI] [PubMed] [Google Scholar]
- 86.Saini V, Farhana A, Glasgow JN, Steyn AJ. Iron sulfur cluster proteins and microbial regulation: Implications for understanding tuberculosis. Curr Opin Chem Biol. 2012;16:45–53. doi: 10.1016/j.cbpa.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Joslin SN, Hendrixson DR. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J Bacteriol. 2008;190:2422–2433. doi: 10.1128/JB.01827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mba Medie F, Champion MM, Williams EA, Champion PAD. Homeostasis of N-α-terminal acetylation of EsxA correlates with virulence in Mycobacterium marinum. Infect Immun. 2014;82:4572–4586. doi: 10.1128/IAI.02153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: Linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 90.Baker JJ, Johnson BK, Abramovitch RB. Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources. Mol Microbiol. 2014;94:56–69. doi: 10.1111/mmi.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson BK, Scholz MB, Teal TK, Abramovitch RB. SPARTA: Simple program for automated reference-based bacterial RNA-seq transcriptome analysis. BMC Bioinformatics. 2016;17:66. doi: 10.1186/s12859-016-0923-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.