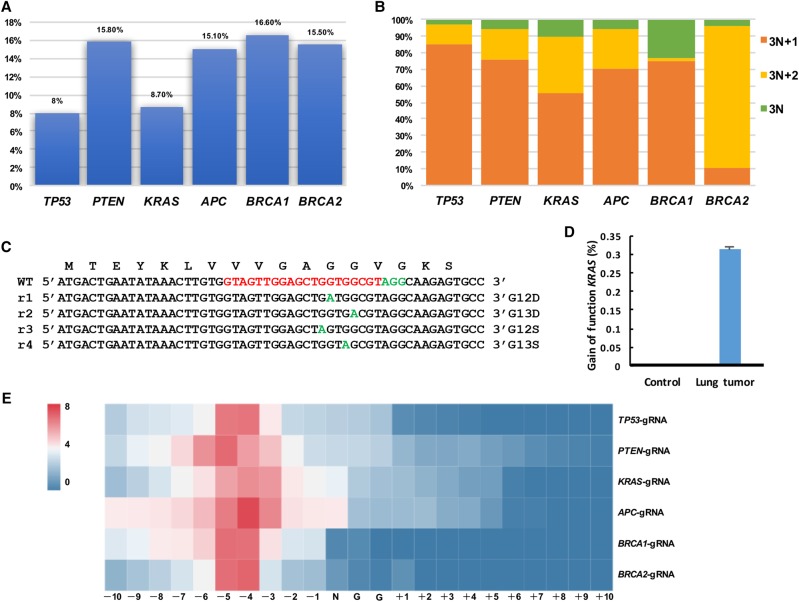
Figure 6.
Mutation analysis in autochthonous lung tumors. (A) Efficiency of indel mutations in sectioned lung tumors was analyzed by deep sequencing. All six sgRNAs could induce indel mutations near the predicted cleavage sites (8.0% of TP53, 15.80% of PTEN, 8.70% of KRAS, 15.10% of APC, 16.60% of BRCA1, and 15.50% of BRCA2). (B) Calculation of mutation patterns (3N, 3N + 1, and 3N + 2) in sectioned lung tumors. (C,D) Gain-of-function mutations of KRAS in tumor cells. (E) Heat map analysis of the mutation efficiency at each position (−10 bp–+10 bp) around PAM sites with different sgRNAs.