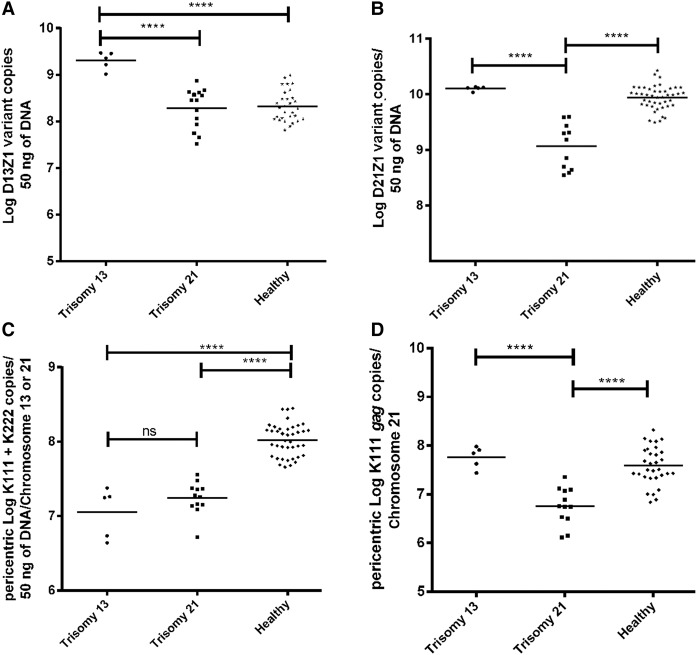
Figure 4.
Centromere and pericentromere instability in individuals with trisomy 21. Detection of D13Z1 (A) and D21Z1 (B) variants in individuals with trisomy 13 or 21 by qPCR using LNA primers and clamps. The copy number of each repeat variant was determined in 50 ng of DNA. (C) Detection of K111 and K222 provirus sequences used as markers to study human pericentromeres. We have developed a PCR assay for K111 plus K222 env (C) and a PCR assay specific for K111 gag (D), in order to assess the structural variation (length) of pericentromeres 13 and 21 in DNA from healthy individuals and individuals with trisomy 13 or 21. The K111 + K222 assay (C) can predict the length of pericentromeres 13 and 21, whereas the K111-specific PCR (D) can predict the length mostly of pericentromere 21. In contrast to healthy individuals, loss of pericentromeric K111 sequences was seen in the DNA of individuals with trisomy 21. Statistical significance among the groups was calculated using the t-test. (****) P-values <0.0001 are shown.