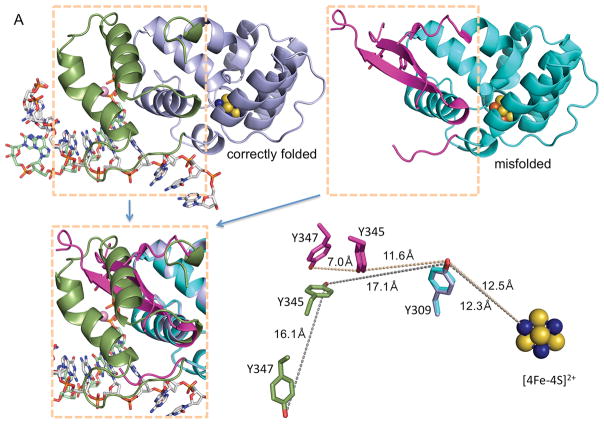
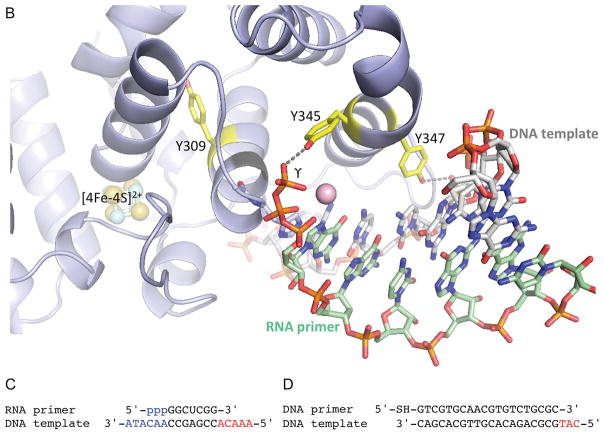
Fig. 1. Comparison of functionally relevant primase substrate and correctly folded p58C (4) with the substrate and misfolded p58C used by O’Brien et al. (1).
(A) Side-by-side comparison of p58C structures with correctly folded (PDB code 5F0Q) and misfolded (PDB code 3L9Q) substrate-binding regions highlighted in different colors. These regions are overlapped in the left bottom quarter. The positions of Y309, Y345 and Y347 relative to [4Fe-4S]2+ in two structures are shown in the right bottom quarter. The reason for p58C misfolding might be explained by substitution of structurally important I271 by serine in erroneously named “wild-type” p58C (PDB code 3L9Q) or by asparagine in its mutants Y345F and Y347F (PDB codes 5I7M and 5DQO, respectively). (B) Crystal structure of p58C in complex with a duplex containing 5′-triphosphate RNA and 3′-overhang DNA (4). (C) Primase substrate (6) with the 5′-triphosphate of a primer and the 3′-overhang of a template that are required for p58C binding are shown in blue. (D) Substrate used by O’Brien et al. (1) for electrochemistry experiments. In (C) and (D) the red-colored 5′-overhangs do not participate in binding of correctly folded p58C. The figure was prepared using the PyMOL Molecular Graphics System (version 1.8, Schrödinger, LLC).