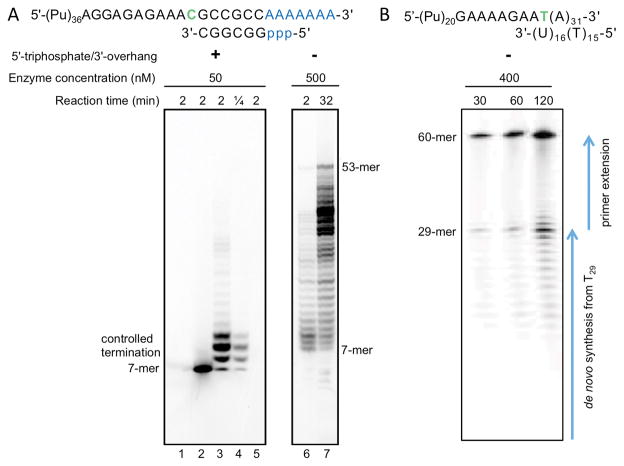
Fig. 2. Side-by-side comparison of experimental setups for examination of RNA synthesis termination by human primase.
(A) Example of primer elongation reactions showing the effect of template:primer structure on the efficiency of RNA synthesis termination by human primase as described by Baranovskiy et al. (6). When using the correct substrate containing 5′-triphosphate and 3′-overhang, there is pronounced termination of reaction mainly at 9-mer primers (lanes 3 and 4), which are optimal for extension by Polα (4). However, in the case of template:primer without 5′-triphosphate and 3′-overhang, primase loses the ability to terminate synthesis at 9-mer primers (lanes 6 and 7) and has dramatically reduced activity, which requires a higher load of the enzyme and longer reaction time. Primase activity reactions were reproduced as in (6). The products were labeled by incorporation of [α-33P]GTP at the seventh position of primer. Note that the negatively charged triphosphate moiety increases the mobility of RNA primers. Lanes 1 and 5, control incubations in absence of enzyme or primer, respectively. Lane 2, reaction was not supplied with CTP and UTP. (B) Example of incorrectly designed primer elongation reactions, which are not capable of capturing physiologically relevant primer termination. The image was adopted from Fig. 5A (1). Note high enzyme concentrations and long reaction time. The products with a length of 29 nucleotides or less are the result of RNA synthesis initiated from T29 in the template (primase initiates RNA synthesis only from a template pyrimidine in the presence of ATP or GTP (13)). Appearance of primers longer than 10 nucleotides is due to the absence of Polα or its catalytic core, which allows primase to rebind the 9-mer primer without involvement of p58C and extend further (6, 14). Unfortunately, the gel provided does not allow visualization of the products of de novo synthesis from 2-mer to 9 to 12-mer. Counting of bands below 29-mer product indicates that the lowest visible band corresponds to 18-mer primer, not to 10-mer as shown in Fig. S14 in (1).