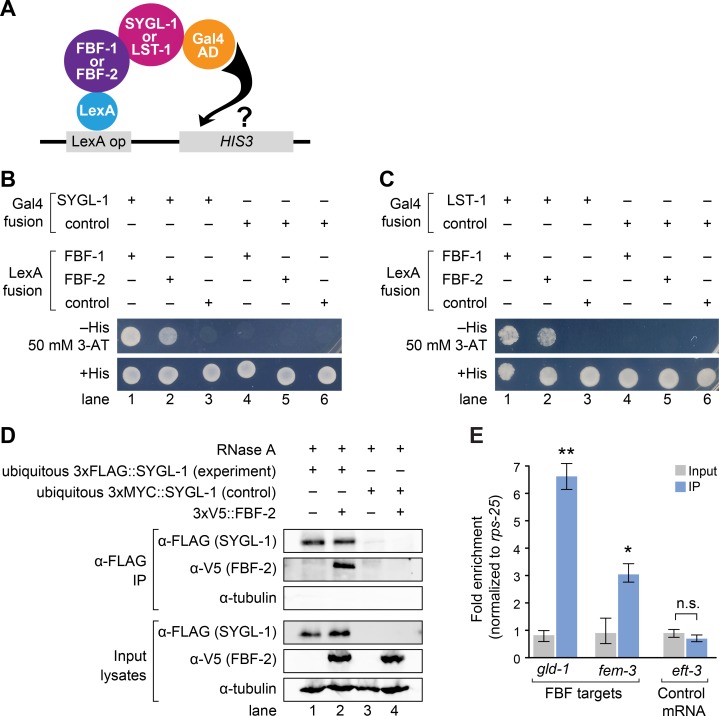
Fig 5. SYGL-1 and LST-1 interact physically with FBF.
(A) Yeast two hybrid assay. Full length SYGL-1 or LST-1 was fused to Gal4 activation domain (AD); PUF repeats of FBF-1(121–614) or FBF-2(121–632) were fused to LexA binding domain (BD). Interaction activates transcription of HIS3 gene. (B and C) Yeast growth assays tested interaction between SYGL-1 and FBF (B) or LST-1 and FBF (C). Yeast strains were monitored for growth on synthetic defined media (SD), either lacking histidine or with histidine as a control. A HIS3 competitive inhibitor (3-AT) improved stringency. (D) SYGL-1 and FBF-2 co-immunoprecipitation (IP). Western blots probed with α-FLAG to detect SYGL-1, α-V5 to detect FBF-2, and anti-α-tubulin as a loading control. 2% of input lysates and 20% of IP elutes were loaded. Exposure times of input and IP lanes are different, so band intensities are not comparable. RNA degradation by RNase A was confirmed. Genotypes for each lane: (1) sygl-1(tm5040); qSi235[Pmex-5::3xFLAG::sygl-1::tbb-2 3’end]; (2) sygl-1(tm5040); fbf-2(q931)[3xV5::fbf-2] qSi235[Pmex-5::3xFLAG::sygl-1::tbb-2 3’end]; (3) sygl-1(tm5040); qSi297[Pmex-5::3xMYC::sygl-1::tbb-2 3’end]; (4) sygl-1(tm5040); fbf-2(q932)[3xV5::fbf-2] qSi297[Pmex-5::3xMYC::sygl-1::tbb-2 3’end]. See S7 Fig for data supporting functionality of epitope-tagged FBF-2. (E) Quantitative PCR of two signature FBF target mRNAs and a control mRNA after α-FLAG IP, using either 3xFLAG::sygl-1(ubiq) for the experiment or 3xMYC::sygl-1(ubiq) as the control. Abundance of mRNAs in input (gray bars) and IPs (blue bars) was calculated with the ΔΔ CT method, using rps-25 for normalization. Error bar indicates standard error. Asterisks indicate a statistically significant difference by 1-way ANOVA with Tukey HSD post hoc test. * p<0.05, ** p<0.01.