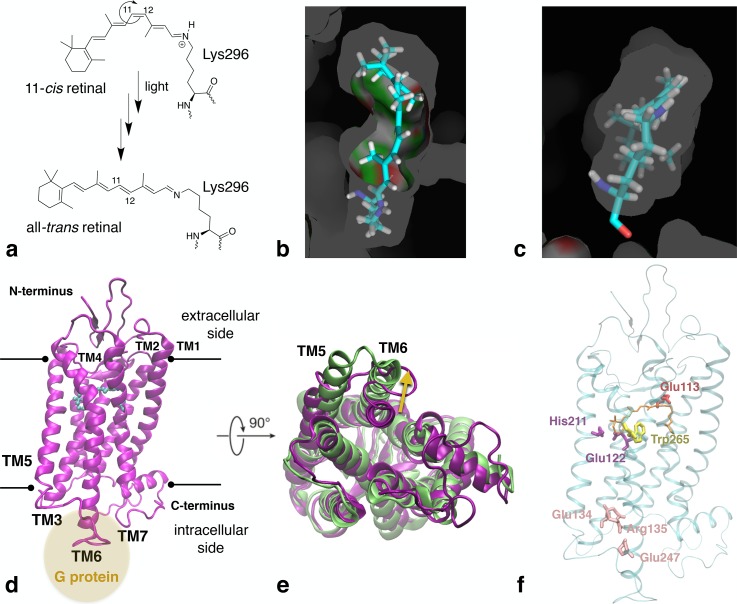
FIG. 15.
Rhodopsin's activation mechanism. (a) 11-cis to all-trans photoisomerisation and later deprotonation of covalently bound retinal in rhodopsin due to exposure to light. (b) View from the extracellular region of the retinal moiety in the active site. (c) View of the retinal moiety, turned 90° compared to image (b), with the Lys296 part located in front. (d) X-ray structure of inactive rhodopsin (PDB code 1U19). The black lines indicate the location of the cell membrane. The yellow circle shows the location of the G-protein interaction site. (e) Alignment of the active and inactive conformation of rhodopsin, viewed from the intracellular side. Corresponding PDB codes are listed in the same colour as the colour of the protein structure. The main structural changes due to activation are indicated by yellow arrows. (f) X-ray structure of inactive rhodopsin (PDB code 1U19) in which specific regions that are especially affected by retinal's cis-trans isomerisation are highlighted. The retinal moiety is shown in orange.