Fig. 3.
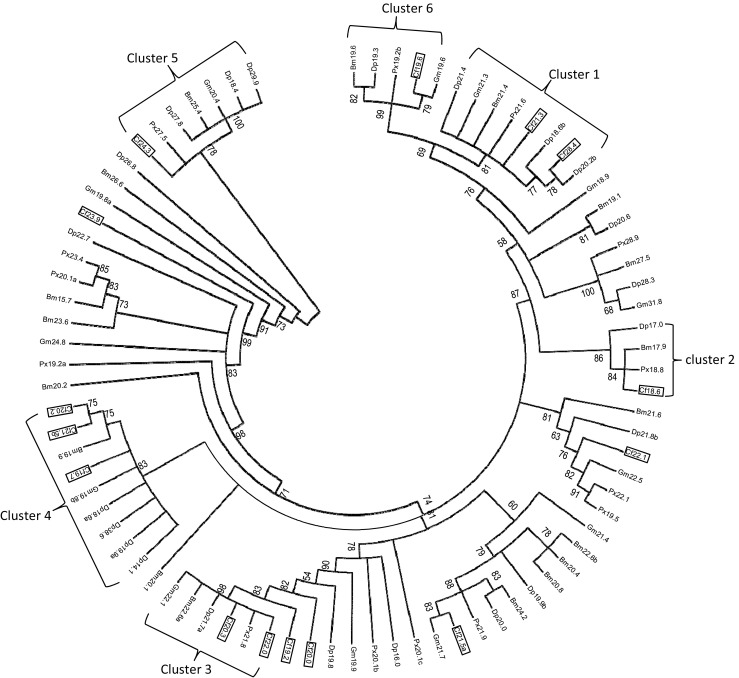
Phylogenetic tree of lepidopteran sHSPs. To construct this phylogenetic tree, 82 lepidopteran sHSP genes were used from the following species: silkworm, B. mori (18 sHSPs); the monarch butterfly, D. plexippus (22 sHSPs); the oriental fruit moth, G. molesta (13 sHSPs); the diamondback moth, P. xylosyella (14 sHSPs); and spruce budworm, C. fumiferana (15 sHSPs). The GenBank accession numbers and ACD domains used for construction of the tree are listed in S1 (Table 1) and S2 (Fig. 1). Only the conserved α-crystallin domain in each sHSP was used in alignment and tree construction. The maximum-likelihood (ML) phylogenetic tree was inferred under the best-fit model LG + G, which was estimated by ProTest. The bootstrap values from 100 resamplings are shown at each node. Branches corresponding to partitions reproduced in less than 50% of the bootstrap replicates were collapsed. Abbreviations of species names: Bm Bombyx mori, Cf Choristoneura fumiferana, Dp Danaus plexippus, Gm Grapholita molesta, Px Plutella xylostella
