Fig. 1.
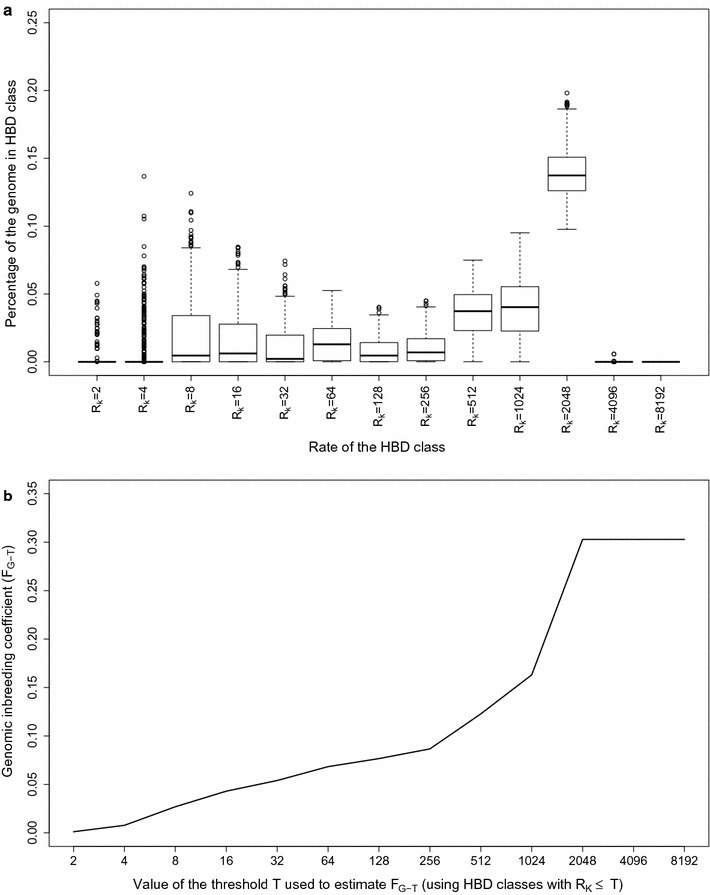
Partitioning of genome-wide autozygosity for the 634 Belgian Blue sires using the BovineHD SNP panel. a Boxplot of percentages of individual genomes associated with 13 HBD-classes with pre-defined R k rates (Mix14R model). The percentages correspond to individual genome-wide probabilities of belonging to each of the HBD-classes. b Genomic inbreeding coefficients estimated with respect to different base populations (F G-T) obtained by selecting different thresholds T that determine which HBD-classes are considered in the estimation of F G-T (e.g., setting the base population approximately 0.5 * T generations in the past). The corresponding inbreeding coefficients F G-T are estimated as the probability of belonging to any of the HBD classes with a R k ≤ T averaged over the whole genome
