Fig. 2.
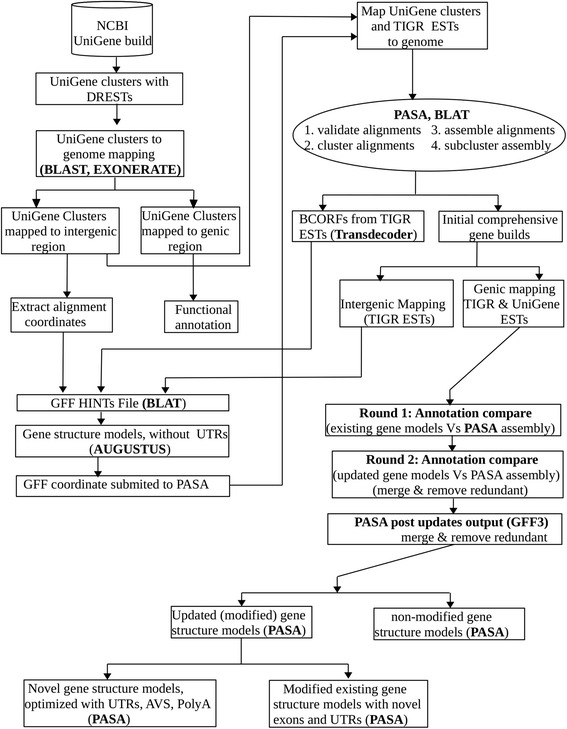
Pipeline for mapping experimental data to reference genome and annotation comparison. This pipeline represents a work flow for identifying known and novel candidate drought responsive genes (CDRGs) and for finding out annotation updates. Identified known putatively uncharacterised genes were functionally annotated. The UniGenes that mapped to integenic region were used by BLAT to generate HINTs and then by AUGUSTUS to identify novel genes which were further optimized by PASA. The PASA pipeline was initiated afresh by cleaning up of any existing output in the MYSQL database using utility codes. The process for annotation comparison was then started by running alignment assembly and by employing the minimum criteria for overlapping transcript alignments and for sub clustering into gene structure (Table 4). Mapping valid alignment assemblies to genome resulted established ICGBs. While the gene builds mapped to the intergenic region that come from the TIGR transcripts were used by BLAT to generate additional HINTs, those mapped to the genic region were used for further annotation comparison. A two round approach was implemented by PASA for processing a complete annotation comparisons: 1st, compared existing gene structure annotations with alignment assemblies and 2nd, re-run, using the output from the first round to capture a few more updates or to verify the initial updates if there was no further updates from the second round. Analysis of alternative spliced alignments and identification of BCORFs were also included in the process. The BCORFs originated from TIGR ESTs were another input to generate HINTs
