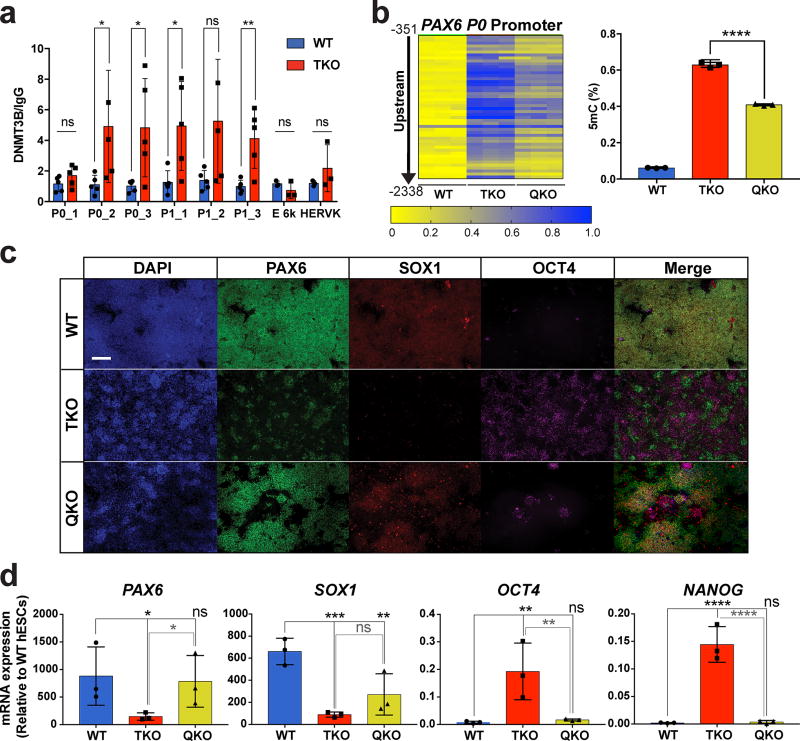
Figure 6. Genetic inactivation of DNMT3B partially rescues the neuroectoderm differentiation defect of TKO hESCs.
a, ChIP-qPCR for DNMT3B at the PAX6 locus in WT and TKO hESCs, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), *P<0.05, **P<0.01. b, Heat map of MassArray analysis of 5mC at the PAX6 P0 promoter in WT, TKO and QKO hESCs. WT and TKO hESCs are passage-matched with the QKO hESCs. The location of each row of CpGs with respect to the TSS is shown to the left of the heat map. For each cell line three independent experiments are shown as three columns. Quantification of the percent methylation is shown on the right, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: Student’s t test (two sided), ****P<0.0001. c, Immunofluorescence of PAX6, SOX1 and OCT4 at D10 of NE differentiation in WT, TKO and QKO cells. Scale bar indicates 100 µm. d, qPCR analysis of neuroectoderm (PAX6 and SOX1) and pluripotency (OCT4 and NANOG) markers in WT, TKO and QKO cells at D10 of NE differentiation, n= 3 independent experiments. Data presented are mean ± STD. Statistical analysis: black lines indicate comparisons to WT, one-way ANOVA, *P<0.05,**P<0.01,***P<0.001, ****P<0.0001.