Abstract
Vascular cell adhesion molecule 1 (VCAM-1) is an important inflammatory biomarker correlating with retinal disease progression. Thus, detection of VCAM-1 mRNA expression levels at an early disease stage could be an important predictive biomarker to assess the risk of disease progression and monitoring treatment response. We have developed VCAM-1 targeted antisense hairpin DNA-functionalized gold nanoparticles (AS-VCAM-1 hAuNP) for the real time detection of VCAM-1 mRNA expression levels in retinal endothelial cells. The AS-VCAM-1 hAuNP fluorescence enhancement clearly visualized the TNF-α induced cellular VCAM-1 mRNA levels with high signal to noise ratios compared to normal serum treated cells. The scrambled hAuNP probes were minimally detectable under same image acquisition conditions. Intracellular hAuNPs were detected using transmission electron microscopy (TEM) analysis of the intact cells. In addition, the AS-VCAM-1 hAuNP probes exhibited no acute toxicity to the retinal microvascular endothelial cells as measured by live-dead assay.
Keywords: VCAM-1, mRNA, microvascular endothelial cells, imaging, gold nanoparticle
Graphical Abstract
This manuscript describes a new nanotechnology for real-time imaging of an inflammatory biomarker in activated retinal microvascular endothelial cells (RMEC). Vascular inflammation is a common component of many retinopathies including diabetic retinopathy (DR) and age-related macular degeneration (AMD). We demonstrate that tumor necrosis factor alpha (TNF-α) induces vascular cell adhesion molecule-1 (VCAM-1) mRNA, a putative vascular inflammatory biomarker, in mouse RMEC. In addition, we have developed novel hairpin-DNA functionalized gold nanoparticles and used them to detect VCAM-1 mRNA in living, TNF-α activated MRMECs. Moreover, hAuNP had no effect on cell viability, indicating that hAuNPs are not acutely toxic to MRMECs. These findings may provide a framework for a new strategy to detect and monitor retinal inflammatory diseases.
BACKGROUND
Molecular biomarkers of retinal inflammation may be exploited to detect and monitor the early onset and progression of associated retinal pathologies (1, 2). Elevated levels of vascular cell adhesion molecule 1 (VCAM-1) have been reported in a number of human retinal diseases (3–5) and in correlated experimental disease models (6, 7). Additionally, it is induced in cultured cells and tissues by the pro-inflammatory cytokine tumour necrosis factor-α (TNF-α) (8). Therefore, VCAM-1 may represent a biomarker of retinal disease, and its in vivo spatiotemporal imaging could be applicable to disease staging. Several attempts have been made to visualize VCAM-1 protein levels with radiolabeled antibodies; however, these methods have been limited by low target-to-background ratios (9, 10). Development of nanomaterial-based probes targeting VCAM-1 protein have overcome some of these limitations, but problems still exist with their application to in vivo imaging (11–13). Thus, an alternative approach may be to image VCAM-1 mRNA in cells and tissues; and if successful, this novel method could potentially provide increased specificity and utility.
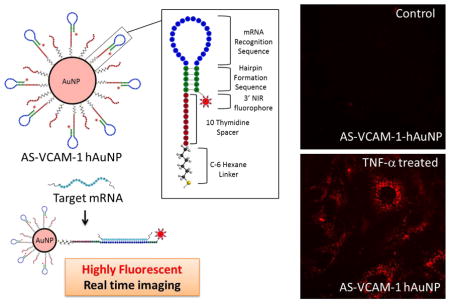
Both others and we have explored the application of hairpin DNA-functionalized gold nanoparticles (hAuNP) to image intracellular mRNAs in living cells (14–16). The DNA hairpin consists of an oligonucleotide modified with a fluorophore on its 3′ end; it incorporates an anti-sense sequence targeting an mRNA of interest, and a self- complimentary sequence that maintains the hairpin structure. The 5′ end is modified with a thiol (-SH) group and it forms a strong Au-S bond that anchors the DNA hairpin to the surface of the gold nanoparticle. The strategy for the application of hAuNP to in vivo imaging relies in part on the quenching properties of gold. In the absence of mRNA target sequence, evidence indicates that hAuNP maintain the hairpin structure, and this structure requires the fluorophore to be juxtaposed to the surface of the gold nanaoparticle. In this configuration, the gold-dependent quenching mechanism is optimal. However, in the presence of target mRNA, hybridization to the anti-sense sequence causes the hairpin to linearize, displacing the fluorophore to distances beyond those required for quenching, and inducing the mRNA-hybridized hAuNP to become highly fluorescent. We have previously reported the advantages of hAuNP and their application to imaging mRNAs in living cancer cells (16, 17). In the current study, we have constructed hAuNP functionalized with DNA hairpins incorporating anti-sense sequence complimentary to VCAM-1 mRNA (AS-VCAM-1 hAuNP). We tested this probe in TNF-α-activated mouse retinal microvascular endothelial cells (MRMECs) to determine if it could specifically image increased intracellular VCAM-1 mRNA. Herein, we report the results.
METHODS
Materials
All chemicals were purchased form Sigma-Aldrich (St. Louis, MO) and used as received unless otherwise noted. The mouse primary retinal microvascular endothelial cells (MRMEC) were obtained from Cell Biologics, Inc. (Chicago, IL). Gold nanoparticles with diameters ranging from 15–20 nm were obtained from Nanopartz, Inc. (Loveland, CO). Custom designed molecular beacons (MB) and custom oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA).
Design and synthesis of hAuNP
DNA oligonucleotides were synthesized that incorporated the mouse anti-sense VCAM-1 sequence GCC TCC ACC AGA CTG TAC GAT CCT; hairpin formation sequence 5′-GC AGC GCC TCC ACC AGA CTG TAC GAT CCT GCT GC-3′ (complementary to AGG ATC GTA CAG TCT GGT GGA GGC) or a scrambled version of this sequence (nonsense sequence) ATA ACT CGT CCG TCC GTA CCG ACC; hairpin formation sequence 5′-GC AGC ATA ACT CGT CCG TCC GTA CCG ACC GCT GC-3′ (complementary to GGT CGG TAC GGA CGG ACG AGT TAT). The anti-sense VCAM-1 sequence was extensively BLAST searched (e-value of 7e-05) to confirm no significant overlap with any other mouse mRNA transcript. The same was performed on the non-sense sequence to confirm non-recognition of any transcribed mouse sequence. The anti-sense VCAM-1 and the nonsense sequences are located within the loop of the hairpin structure (Figure 1A). A self-complementary sequence was incorporated into the DNA oligonucleotides, forming the stem of the DNA hairpin (Figure 1A). This sequence is largely responsible for the formation and the stability of the hairpin secondary structure. Each DNA-oligonucleotide was computationally designed via energy minimalization to achieve the formation of the hairpin structure (Figure 1A). Each of the optimized DNA-oligonucleotide strands was coupled to an Alexafluor-647 near-infrared (NIR) dye (fluorophore) at the 3′ end. The 5′ end was modified with a thiol group to facilitate linkage to the surface of the gold nanoparticles via an Au-S bond. The hAuNPs were synthesized according to our previously described method (16). Prior to use, thiol-terminated oligonucleotides were subjected to 0.1 M dithiolthreitol (DTT) reduction of the 5′ thiol moiety. Excess reducing agent was removed by centrifugal filtration using a filter with a 3K molecular weight cut-off (Amicon Ultracel 3K from Millipore; Billerica, MA). The freshly activated 5′ thiol-modified oligonucleotide strands were washed three times with PBS (Life Technologies Corporation; Grand Island, NY) and stored at −80°C. The average diameter of the hAuNP was determined to be approximately 37±4 nm by dynamic light scattering (DLS) (Figure S1A). The diameters of the gold nanoparticles used to prepare the hAuNP ranged from 15–20 nm by transmission electron microscopy (TEM) analysis (Figure S1B). The number of DNA-oligonucleotide strands per gold nanoparticle was approximately 48, as quantified by fluorescence measurements after digestion of the hAuNP using DTT (Figure S1C), a method described previously (18).
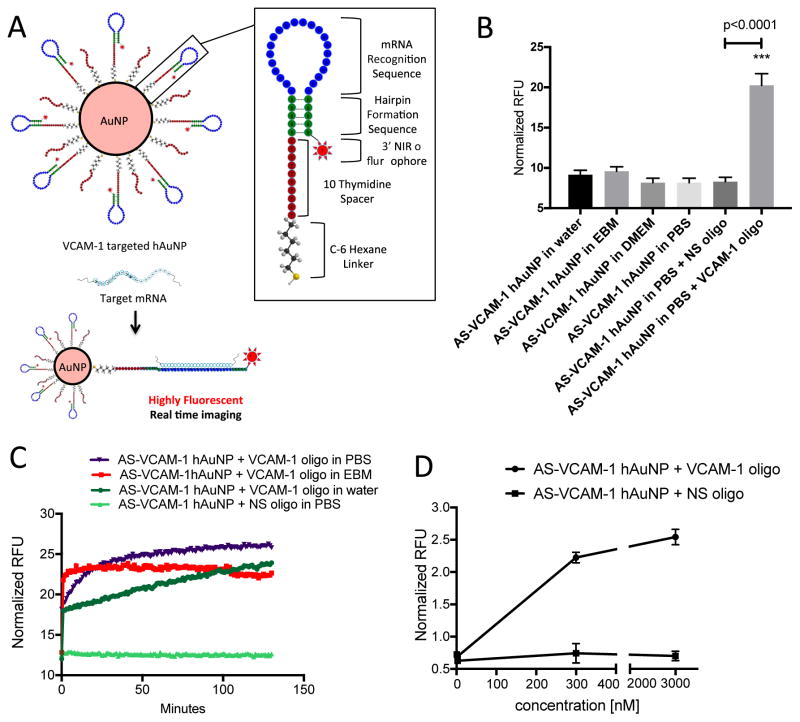
Figure 1.
Structural design, stability, sensitivity and hybridization kinetics of hAuNP. (A) Gold nanoparticles were functionalized with DNA hairpin-loops incorporating anti-sense sequence specific for mouse VCAM-1 mRNA (AS-VCAM-1 hAuNP) or a scrambled version of this sequence (NS hAuNP). DNA hairpin-loops are modified on their 5′ ends with a thiol (SH) group and coupled to gold nanoparticles through an Au-S bond. In the absence of VCAM-1 mRNA, the stem region (green) maintains the hairpin structure through self-complementary hydrogen bonding. The hairpin structure requires the 3′ near-infrared (NIR) fluorophore to be held in close proximity to the gold surface. The capacity of gold to quench fluorescence is maximal in this configuration; therefore, no fluorescence is observed. Hybridization of a target mRNA to the anti-sense recognition sequence (blue) causes the hairpin to open, increasing the distance between the NIR dye and the gold surface, and resulting in fluorescence emission. (B) AS-VCAM-1 hAuNP are stable in a variety of media. Fluorescence was only observed when a complimentary single strand (ss) DNA oligonucleotide (VCAM-1 oligonucleotide) was present, indicating the specificity of the AS-VCAM-1 hAuNP. (C) The hybridization kinetics of the AS-VCAM-1 hAuNP in the presence of the VCAM-1 oligonucleotide depended on the reaction medium. These studies also confirmed the specificity of the AS-VCAM-1 hAuNP for the exogenous VCAM-1 oligonucleotide compared to the scrambled sequence (NS oligonucleotide). (D) AS-VCAM-1 hAuNP exposed to increasing doses of the VCAM-1 oligonucleotide exhibited a dose-dependent increase in fluorescence. The same effect was not observed when AS-VCAM-1 hAuNP was exposed to the NS oligonucleotide.
Dynamic Light Scattering (DLS)
All DLS measurements were performed on a Malvern Zetasizer Nano ZS (Malvern Instruments, Inc.; Westborough, MA). Particle measurements were performed at a concentration of 1 nM hAuNP in PBS (Life Technologies Corp.; Carlsbad, CA). These measurements were performed in triplicate.
Cell Culture
MRMECs were cultured in T-75 cell culture flasks (Thermo Fisher Scientific; Wilmington, MA) coated with attachment factor (Cell Systems; Danvers, MA) and in growth medium consisting of endothelial basal medium (EBM; Lonza; Walkersville, MD) supplemented with 2% FBS (Lonza) and endothelial cell growth supplements (EGM SingleQuots; Lonza). All cultures were incubated at 37°C, 5% CO2 and 95% relative humidity (20.9% oxygen). The cells were cultured in four-well chamber slides and treated with 0.5 nM hAuNP in complete growth medium. Passages 4 to 6 were used in these experiments. Each experiment was repeated at least three times.
Anti-sense VCAM-1 hAuNP target specificity
To determine target specificity, anti-sense VCAM-1 hAuNP (1 nM, PBS, 0.1% tween-20) were titrated with increasing concentrations of VCAM-1 recognition complement (VCAM-1 oligonucleotide strand) (0–1 μM) and scrambled complement (NS- oligonucleotide strand) (0–1 μM). The hybridization solutions were sealed, protected from light and incubated for 2 hrs at 37°C. Fluorescence intensities were measured using a microplate reader (Biotek Instruments, Inc.; Winooski, VT) and plotted as a function of complement concentration. Signal to noise was determined by the fluorescence ratio of the VCAM-1 vs the NS-oligonucleotide strand. Experiments were performed at least three times with n=3 for each experimental group.
qRT-PCR
VCAM-1 mRNA expression levels in MRMEC cells were determined by qRT-PCR. MRMECs were seeded in 4-well chamber slides at 2 × 104 cells/chamber and cultured under growth conditions. At 75% confluence, MRMECs were treated with complete medium plus 10 ng/ml TNF-α (Millipore; Billerica, MA) or vehicle (PBS, Life Technologies) for 6 hrs. After treatment, cells were washed with cold PBS (Life Technologies) and total RNA was isolated using an RNeasy kit (Qiagen; Germantown, MD), according to the manufacturer’s instructions. Total RNA was reverse transcribed using the High-Capacity cDNA Archive Kit (Life Technologies). qRT-PCR was performed by co-amplification of the VCAM1 gene vs b-actin (endogenous control) using gene-specific TaqMan Gene Expression Assays (Gene Symbol: Vcam1, mCG19764; assay ID: Mm01320970_m1; b-actin: Gene Symbol: Actb, mCG23209; Assay ID: Mm00607939_s1 from Life Technologies). Expression data were analyzed using the comparative Ct method. Experiments were performed at least three times with three replicates for each experimental group.
Confocal Microscopy of hAuNP
MRMECs (2 X 104) were cultured in Lab-Tec II chamber slides (Sigma Aldrich; St. Louis, MO) and in complete EBM growth medium including growth supplements and 2% FBS, all from Lonza, supplemented with 0.5 nM AS-VCAM-1 or NS hAuNP. These cultures were treated with TNF-α (10 ng/ml) or vehicle for 6 hrs. The media with their respective treatments were aspirated and the cells were washed three times with PBS (Life Technologies). Live cell imaging was performed using a Nikon spinning disk high-speed live cell confocal imaging microscope (Melville, NY) configured for hAuNP-specific emission. DAPI (Fisher Scientific Company LLC, Suwanee, GA) was used as a nuclear counterstain according to the manufacturer’s instructions. Cell viability was monitored in cultured cells processed in parallel using trypan blue (Fisher Scientific Company LLC) to ensure >95% viability throughout imaging experiments. Cells were imaged using Nikon confocal microscope (Nikon, Inc.; Melville, NY). Experiments were performed at least three times with three replicates for each experimental group.
Transmission Electron Microscopy (TEM)
Embedding
After incubation with hAuNPs, MRMECs were fixed in 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer, pH 7.4 at room temperature (RT) for 1 hour and then transferred to 4°C, overnight. The cells were washed in 0.1M sodium cacodylate buffer and incubated for 1 hour in 1% osmium tetraoxide at RT, then washed with 0.1M sodium cacodylate buffer. The cells were dehydrated through a graded ethanol series followed by three exchanges of 100% ethanol. Next, the cells were incubated for 5 minutes in 100% ethanol and propylene oxide (PO) followed by two exchanges of pure PO. Samples were infiltrated with 25% Epon 812 resin and 75% PO for 30 minutes at RT, then overnight with Epon 812 resin and PO [1:1] at RT. The next day, the Epon 812 resin and PO [1:1] was exchanged for 3–4 hours in resin and PO [3:1], then incubated with pure epoxy resin overnight. The cells were then incubated in two more changes of pure epoxy resin then allowed to polymerize at 60°C for 48 hours.
Sectioning and Imaging
Ultra-thin 70–80-nm sections were cut and collected on 300-mesh copper grids, post-stained with 2% uranyl acetate and then with Reynold’s lead citrate. Samples were subsequently imaged on the Philips/FEI Tecnai T12 electron microscope (Hillsboro, OR) at various magnifications.
Cell viability assay
An EZViable Calcein AM Fluorometric Cell Viability Assay Kit (BioVision; Milpitas, CA) was used to quantify the number of viable cells. MRMECs were cultured on sterile black 96-well plates under growth conditions. At 75% confluence, MRMECs were treated with 0 to 5 nM hAuNP in complete medium, citrate capped GNP (5 nM) in complete medium, or 70% ethanol as a positive control for 8 hrs. After treatment, the cells were washed with cold PBS (Life Technologies). MRMECs were then exposed to a buffered (1:500) calcein AM solution and incubated at 37°C for 30 minutes. Fluorometric readings were performed using a microplate reader (Biotek). Fluorescence intensity was plotted on the Y-axis and represented as % live cells. Experiments were performed at least three times with three replicates for each experimental group.
Statistics
Data were expressed as mean ± SDM and statistical differences among groups were determined by one-way analysis of variance (ANOVA) using Prism 6 (Graph-Pad; San Diego, CA) followed by Bonferroni post hoc test to determine significant differences between specific groups. A p-value <0.05 was considered statistically significant.
RESULTS
Specificity of AS-VCAM-1 hAuNP
AS-VCAM-1 hAuNP and NS hAuNP were prepared and characterized as described in materials and methods and illustrated in Figure 1A. AS-VCAM-1 hAuNP were incubated with an exogenous oligonucleotide strand incorporating VCAM-1 target sequence (VCAM-1 oligonucleotide) at concentrations ranging from 3 to 3000 nM. An approximate 3.5-fold dose-dependent increase in fluorescence activity was observed over this concentration range that was statistically significant (p<0.05) (Figure 1D). There were no statistically significant differences in fluorescence activity when AS-VCAM-1 hAuNP were incubated with an exogenous oligonucleotide incorporating the scrambled VCAM-1 sequence (NS oligonucleotide). AS-VCAM-1 hAuNP were incubated in various media and increased fluorescence activity was observed only in the presence of the VCAM-1 oligonucleotide (Figure 1B). Rates of AS-VCAM-1 hAuNP hybridization with the VCAM-1 oligonucleotide depended on the reaction medium; they were slowest in water, increasing in PBS, and fastest in EBM medium (Figure 1C). This is consistent with the concept that, media with high ionic concentration can accelerate molecular beacon hybridization kinetics. The double strand stem region is relatively rigid (Figure 1A), the probe undergoes a spontaneous conformational changes upon hybridization; however, the rate is limited due to an equilibrium between the original beacon conformation. Kinetics of hybridization was faster in EBM, and the fluorescence intensity reaches to maximum within few minutes. Further coupling reaction time had very little effect on the rate or final fluorescence intensity. The slower hybridization reaction in PBS required longer coupling reaction to reach the final fluorescence intensity which was higher than the hybridization-kinetics in EBM, requiring to be monitored for >2 hours.
Stability of AS-VCAM-1 hAuNP
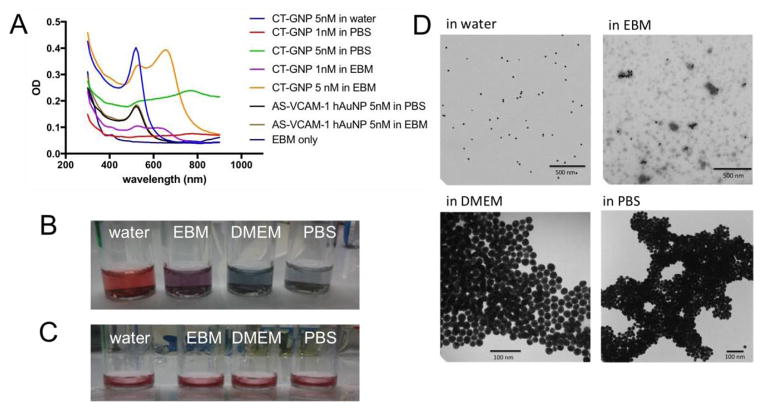
We tested and compared the stabilities of the AS- VCAM-1 hAuNP and citrate capped gold nanoparticles (CT-GNP) in different media (Figure 2). Aggregation of colloidal gold may be detected by changes in absorbance spectra and by TEM. In Figures 2A and 2D, CT-GNP are monodispersed in water; however, as the ionic strength of the aqueous medium is increased, they aggregate as shown in Figure 2D by TEM analysis. Changes observed in their absorbance profiles as shown in Figure 2A also indicate their aggregation (19). An absorbance maximum of 520 nm is observed when CT-GNP are monodispersed in water. In contrast, the absorbance profiles in PBS and EBM become broader, which is characteristic of aggregation. Notably, when colloidal dispersions are prepared from AS-VCAM-1 hAuNP in each of these media, there is little change in the absorbance spectrum, indicating a monodispersion (Figure 2C). In addition, we have previously reported that 15 nm spherical gold-nanoparticles provide superior stability to the oligonucleotides compared to unconjugated oligonucleotide (17).
Figure 2.
Stability of AS-VCAM-1 hAuNP and citrate-capped GNP (CT-GNP) in different media. (A) The spectroscopic analysis of AS-VCAM-1 hAuNP and CT-GNP. The 520 nm absorbance maximum was stable for AS-VCAM-1 hAuNP in PBS and EBM indicating monodispersions. The CT-GNP exhibits an absorbance profile with a maximum 520nm in water. However, in PBS and EBM this profile is significantly altered, presumably due to aggregation. (B) Qualitative analysis of CT-GNP in different media, indicating a monodispersion in water (red). A color shift to blue/violet was observed in isotonic media, thus indicating aggregation of the CT-GNP. (C) Qualitative analysis of AS-VCAM-1 hAuNP indicated their stability (no aggregation) in each of these media (all red). (D) Transmission electron microscopy (TEM) micrograph of CT-GNP showing monodispersion in water, and aggregation in PBS and cell culture medium, consistent with A and B.
Specificity of AS-VCAM-1 hAuNP for mRNA target
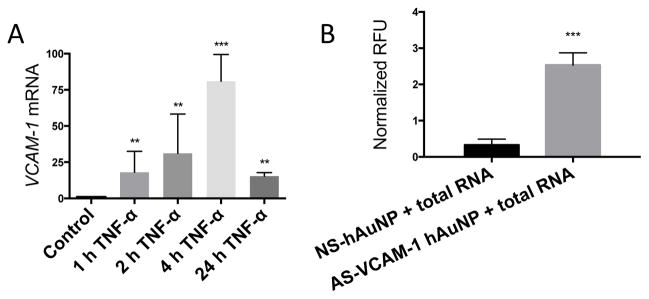
We observed that TNF-α induces expression of VCAM-1 mRNA in MRMEC and peaked at 4 hours by approximately 75-fold over the control, as determined by qRT-PCR (Figure 3A). Thus, we isolated samples of total RNA from MRMEC treated with vehicle or TNF-α for 4 hours and incubated them with AS-VCAM-1 hAuNP or NS hAuNP (Figure 3B). A statistically significant 6.25-fold fluorescence enhancement was observed in hybridization reactions between AS-VCAM-1 hAuNP and mRNA from TNF-α-induced MRMEC (p<0.05). No signal enhancement was observed in the hybridization reactions with NS hAuNP.
Figure 3.
Specificity of AS-VCAM-1 hAuNP for VCAM-1 mRNA. (A) Total RNA was isolated from TNF-α- (10 ng/ml) treated MRMEC at 1 to 24 hours. VCAM-1 mRNA expression levels relative to 18s was determined by qRT-PCR. Expression values are expressed as fold change relative to unstimulated cells. Maximum expression was observed at 4 hours that was increased by approximately 75-fold over the control. (B) Total RNA was isolated from MRMEC treated with TNF-α (10 ng/ml) or vehicle. Hybridization reactions were performed by incubating the total RNA samples with AS-VCAM-1 hAuNP or NS hAuNP at 37°C for 2 hours. A 6.25-fold statistically significant signal enhancement was observed in reactions with AS-VCAM-1 hAuNP compared to those with NS hAuNP. The results of this experiment indicates the ability of AS-VCAM-1 hAuNP to discriminate VCAM-1 mRNA against the TNF-α-induced MRMEC transcriptome.
Internalization of hAuNPs by MRMEC
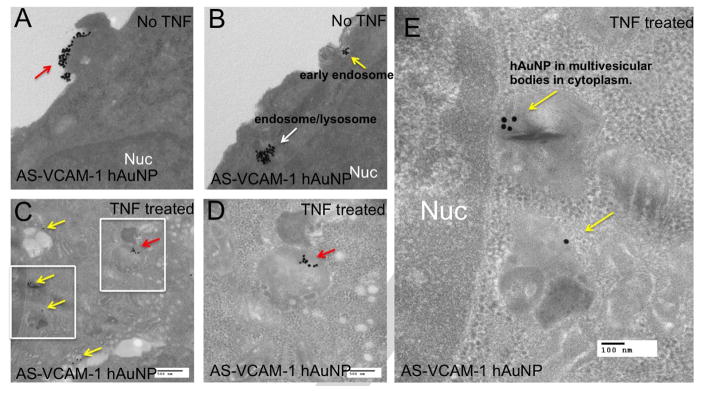
MRMEC were incubated with AS-VCAM-1 or NS hAuNPs, and imaged using TEM (Figure 4 and S2). Independent of their nucleotide sequence, we observed hAuNP in the perinuclear region and throughout the cytoplasm in TNF-α and vehicle-treated MRMEC. We did not observe hAuNP inside the nucleus of MRMECs (Figure 4A vs 4C). A TEM micrograph demonstrates clusters of NS hAuNP localized in either endosomes or lysosomes throughout the cytoplasm of TNF-α treated MRMECs (Figure S2).
Figure 4.
Transmission electron microscopy (TEM) imaging of MRMECs and detection of intracellular AS-VCAM-1 hAuNP. (A and B) TEM micrograph of MRMECs cultured in normal growth medium plus AS-VCAM-1 hAuNP. (A) Clusters of hAuNPs were visible on the surface of the plasma membrane (red arrow). (B) intracellular hAuNP were observed just beneath the plasma membrane (yellow arrow) and further into the cell’s interior (white arrow). (C, D and E) TNF-α treated MRMECs incubated with AS-VCAM-1 hAuNP. (C) Clusters of AS-VCAM-1 hAuNP were localized in endosomes or lysosomes throughout the cytoplasm as indicated by red and yellow arrows. Panel D is an inset from C, the red arrows indicate AS-VCAM-1 hAuNP internalized in a lysosome/endosome. Panel E is another inset from C; the yellow arrows indicate the AS-VCAM-1 hAuNPs internalized in multivesicular bodies in the cytoplasm. AS-VCAM-1- and NS hAuNP were indiscriminately internalized by MRMEC independent of treatment.
Imaging of VCAM-1 mRNA expression levels in living MRMEC
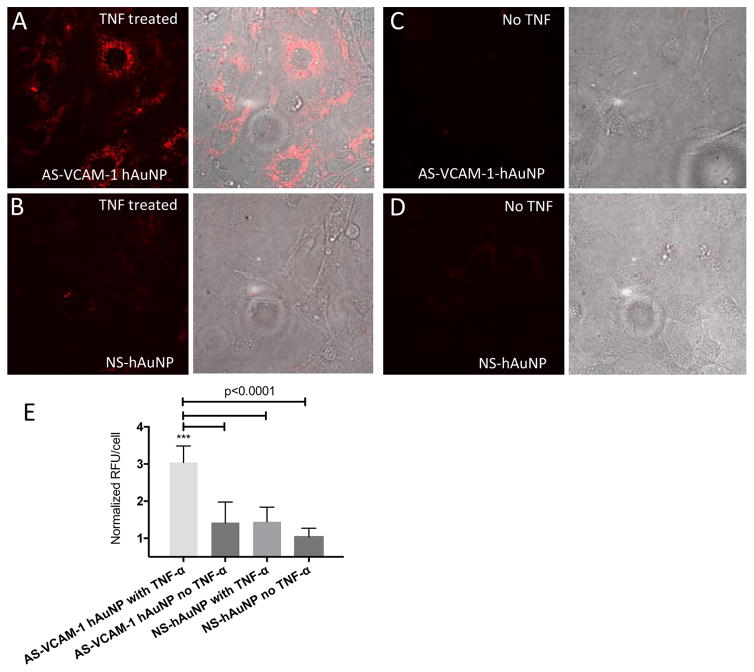
An approximate 2.25-fold increase in AS-VCAM-1 hAuNP-dependent fluorescence enhancement was observed in TNF-α- vs. vehicle-treated MRMECs (Figure 5 and S3). The observed cytoplasmic, perinuclear patches of fluorescence were consistent with the localization of hAuNP determined by our TEM analysis shown in Figure 4. After prolonged incubation, fluorescence was maintained within the intracellular cytoplasmic compartment, supporting intracellular retention (Figure S3). The NS hAuNP probes were minimally detectable in TNF-α- vs. vehicle-treated MRMECs under the same image acquisition conditions. This result is consistent with the in vitro specificity assays shown in Figures 1B and 3B.
Figure 5.
Confocal imaging of live MRMECs treated with AS-VCAM-1 hAuNP and NS hAuNP. Cells were cultured on microscope slides and treated with TNF-α or vehicle plus AS-VCAM-1 hAuNP or NS hAuNP in complete growth medium. After a 6-hour incubation, these media were aspirated and fresh medium was added to each culture. The cells were imaged using confocal microscopy. (A) Strong fluorescence emission was only detected in TNF-α-activated MRMEC treated with AS-VCAM-1 hAuNP. (B, C and D) Only minimal fluorescence was detected in the other cultures. (E) Fluorescence intensity is expressed in relative fluorescence units (RFU) per cell over different treatment groups as measured using ImageJ software.
Cytotoxicity of hAuNP
Cell viability assays were performed in MRMEC treated with variable concentration of AS-VCAM-1 hAuNP or NS hAuNP ranging from 0–5 nM. Calcein AM activation was monitored by fluorescence emission arising from intracellular hydrolysis of the Calcein AM. hAuNP had no effect on cell viability, indicating that hAuNPs are not acutely toxic to MRMEC (Figure 6). Citrate-capped 15nm GNPs reduced cell viability, perhaps due to changes in physical properties and aggregation in cell culture medium as shown in Figure 2.
Figure 6.
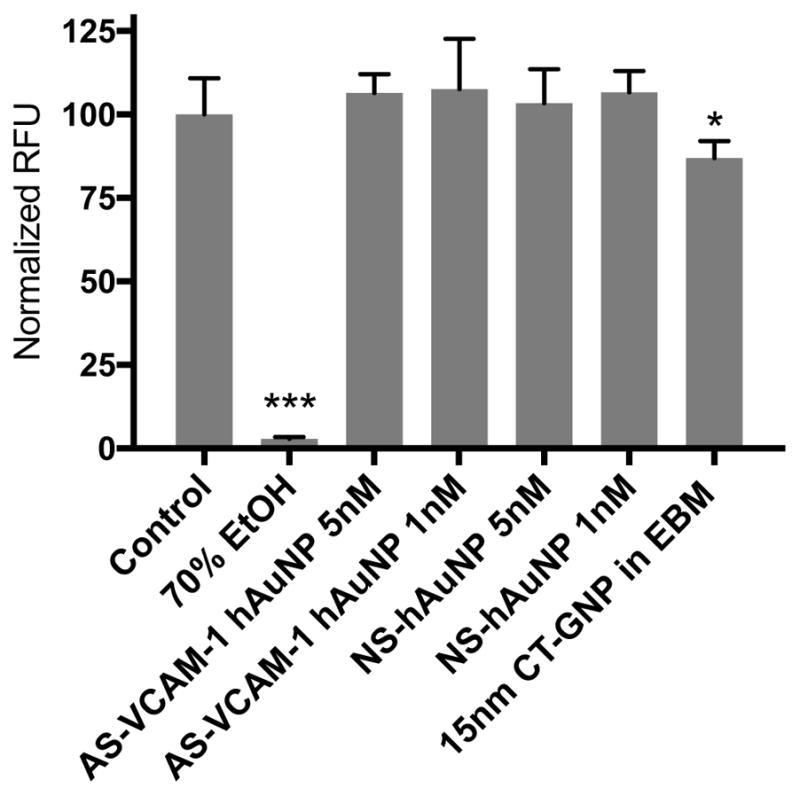
Cytotoxicity of AS-VCAM-1 hAuNP, NS hAuNP and CT-GNP. Cytotoxicity was measured by the live-dead assay using Calcein AM. AS-VCAM-1 and NS hAuNP did not significantly reduce MRMEC viability at 1 and 5 nM when compared to normal serum-treated cells. Citrate-capped 15nm gold nanoparticle (CT-GNP) showed reduced cell viability compared to control (p <0.05). Data are presented as mean ± SD (n = 5).
DISCUSSION
A non-invasive method that detects and images intracellular disease-linked mRNAs would significantly improve the staging of disease progression and allow the assessment of treatment response in the clinic. Gold is inert and “safe” and used successfully in conjunction with a number of clinical therapies and techniques. It is used: as an anti-inflammatory agent to treat rheumatoid arthritis (20), in photothermal cancer therapy, and as an imaging agent (21).
VCAM-1 is a cellular adhesion protein that is induced on the surface of endothelia in vascular inflammation. Diabetic retinopathy (DR) is considered to be a chronic inflammatory condition and evidence has shown that VCAM-1 levels increase in the diabetic retina correlating with disease progression. Through binding its cognate ligand expressed on the surface of leukocytes, VCAM-1 facilitates the firm adhesion of leukocytes (leukostasis) to endothelium in DR, which may trigger a cascade of events that potentially leads to vision loss. TNF-α is an inflammatory cytokine and its serum levels are elevated in diabetics (22). Additionally, it induces the increased expression of VCAM-1 in endothelial cells (23). Guided by these considerations, we chose TNF-α as a disease-relevant inflammatory stimulus and VCAM-1 mRNA as a target for our imaging experiments.
As determined by the absorbance profiles and TEM, monodispersions of AS-VCAM-1 hAuNP are stable in isotonic medium, whereas CT-GNP are not. These data indicate that linking these DNA oligonucleotides to gold nanoparticles confers stability against aggregation. This is a crucial finding, as nanoparticle aggregation in vivo could lead to deleterious side effects precluding their use. TEM showed that AS-VCAM-1 and NS hAuNP are readily internalized by MRMEC, localizing to the cytosol and to perinuclear regions. Some of these were observed presumably in early and late endosomes and/or lysosomes. We did not observe any hAuNP in the nucleus. Gold nanoparticles facilitate cellular internalization while at the same time stabilize the conjugated DNA hairpin against DNase activity; these features are clearly advantageous for in vitro and in vivo applications. The non-specific cellular uptake we observed (Figures 4 and S2), is consistent with previously reported results (16). The hAuNP are indiscriminately internalized by mammalian cells by mechanisms that are not clearly understood (24). Furthermore, uptake mechanisms of hAuNP may vary substantially depending on the nucleotide sequence, the specific configuration of the oligonucleotide, and the overall surface charge.
We demonstrated that AS-VCAM-1 hAuNP specifically hybridized with an exogenous VCAM-1 oligonucleotide in a concentration-dependent manner. Furthermore, the increased hybridization rates observed in media with a physiologically relevant osmolarity are advantageous for in vivo applications. These data suggested that AS- VCAM-1 hAuNP are likely to be specific for VCAM-1 mRNA in the intracellular environment; therefore, we tested their capacity as imaging probes within this context.
Total RNA was isolated from TNF-α-activated MRMEC and hybridization with AS-VCAM-1 hAuNP yielded a 6.25-fold fluorescence enhancement. Minimal background signal was observed from hybridization reactions with NS hAuNP. This experiment demonstrated the capacity of the AS-VCAM-1 hAuNP to discriminate their target against the TNF-α-induced MRMEC transcriptome.
In our in vitro experiments involving the hybridization reactions of AS-VCAM-1 hAuNP with VCAM-1 oligonucleotide or total RNA, we note that the responsivity of the fluorescent signal in comparison to increments in target concentration is low. For example, a 3000-fold increase in VCAM-1 oligonucleotide concentration vs. 3.5-fold fluorescence enhancement, and a 95-fold increase in VCAM-1 mRNA vs 6.25-fold fluorescence enhancement, were observed in these experiments. These low responses are likely to pertain to the basic design of hAuNP. Currently our lab is focusing on redesigning hAuNP for the development of structure vs reactivity profiles to optimize this parameter.
A statistically significant 2.25-fold fluorescence enhancement was observed when TNF-α-activated MRMEC were treated with AS-VCAM-1 or NS hAuNP. These results are comparable to those of a previous study in which hairpin DNA hAuNP were used to distinguish cells that were positive or negative for tyrosinase (17). A 3.5-fold fluorescence enhancement was reported in that study. AS-VCAM-1 or NS hAuNP were non-toxic to MRMEC, although cell death was-induced by citrate capped GNP treatment, as expected (25–27).
Numerous data exist suggesting that the application of hAuNPs to imaging intracellular mRNAs offers significant advantages. For example, the surface functionalization of gold nanoparticles with hairpin DNA-oligonucleotides may enable detection of low abundance intracellular RNAs with high sensitivity. hAuNP are also efficient quenchers of numerous fluorophores (28); therefore, they could foster the development of a methodology for multiplexing the detection and imaging of multiple distinct mRNA targets, using judicious selection of spectrally distinct fluorophores. Continued development of hAuNP-dependent imaging modalities may allow for the collection of quantitative data to discern variable levels of mRNA expression in disease-relevant contexts (29, 30). Ongoing work is directed toward the multiplexed imaging of retinal disease-specific mRNA targets in living animals.
In summary, we have used an AS-VCAM-1 hAuNP probe to detect and image increased VCAM-1 mRNA expression levels, induced by disease relevant stimuli, in living retinal endothelial cells. These probes are readily internalized by cells without the need for transfection reagents or cell permeabilization. These hAuNP imaging probes allowed specific imaging of VCAM-1 mRNA in endothelial cells activated by TNF-α – a cytokine that is associated with the inflammatory component of DR inflammation (24, 31). These findings may provide a framework for a new strategy to detect and monitor retinal inflammatory diseases.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R01EY23397, R01EY07533 and R01EY23639 (to JSP), a grant from the Knights Templar Eye Foundation, Inc. (to MIU), MMPC MICROMouse U24DK076169 (to AJ), a grant from the Carl Marshall Reeves & Mildred Almen Reeves Foundation, Inc. (to JSP), Vanderbilt Diabetes Research and Training Center Core grant (P30 DK020593-34-39), Vanderbilt Vision Research Center NEI Core Grant (P30-EY008126) and an Unrestricted Grant from Research to Prevent Blindness, Inc.
Footnotes
Supporting Information Available
Supplementary Figures are available via the internet.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xie F, Luo W, Zhang Z, Sun D. In vivo molecular imaging in retinal disease. J Ophthalmol. 2012;2012:429387. doi: 10.1155/2012/429387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreuger J, Phillipson M. Targeting vascular and leukocyte communication in angiogenesis, inflammation and fibrosis. Nature Reviews Drug Discovery. 2016;15(2):125–42. doi: 10.1038/nrd.2015.2. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez C, Burgos R, Canton A, Garcia-Arumi J, Segura RM, Simo R. Vitreous levels of vascular cell adhesion molecule and vascular endothelial growth factor in patients with proliferative diabetic retinopathy: a case-control study. Diabetes Care. 2001;24(3):516–21. doi: 10.2337/diacare.24.3.516. [DOI] [PubMed] [Google Scholar]
- 4.Seddon JM, George S, Rosner B, Rifai N. Progression of age-related macular degeneration: prospective assessment of C-reactive protein, interleukin 6, and other cardiovascular biomarkers. Arch Ophthalmol. 2005;123(6):774–82. doi: 10.1001/archopht.123.6.774. [DOI] [PubMed] [Google Scholar]
- 5.Toker E, Kazokoglu H, Sahin S. Cell adhesion molecules in subretinal fluid: Soluble forms of VCAM-1 (vascular cell adhesion molecule-1) and L-selectin. Int Ophthalmol. 1998;22(2):71–6. doi: 10.1023/A:1006160209657. [DOI] [PubMed] [Google Scholar]
- 6.Shen WY, Yu MJT, Barry CJ, Constable IJ, Rakoczy PE. Expression of cell adhesion molecules and vascular endothelial growth factor in experimental choroidal neovascularisation in the rat. Brit J Ophthalmol. 1998;82(9):1063–71. doi: 10.1136/bjo.82.9.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caicedo A, Espinosa-Heidmann DG, Pina Y, Hernandez EP, Cousins SW. Blood-derived macrophages infiltrate the retina and activate Muller glial cells under experimental choroidal neovascularization. Experimental Eye Research. 2005;81(1):38–47. doi: 10.1016/j.exer.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Sawa Y, Sugimoto Y, Ueki T, Ishikawa H, Sato A, Nagato T, Yoshida S. Effects of TNF-alpha on leukocyte adhesion molecule expressions in cultured human lymphatic endothelium. J Histochem Cytochem. 2007;55(7):721–33. doi: 10.1369/jhc.6A7171.2007.. [DOI] [PubMed] [Google Scholar]
- 9.Nahrendorf M, Keliher E, Panizzi P, Zhang H, Hembrador S, Figueiredo JL, Aikawa E, Kelly K, Libby P, Weissleder R. 18F-4V for PET-CT imaging of VCAM-1 expression in atherosclerosis. JACC Cardiovasc Imaging. 2009;2(10):1213–22. doi: 10.1016/j.jcmg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McHale JF, Harari OA, Marshall D, Haskard DO. Vascular endothelial cell expression of ICAM-1 and VCAM-1 at the onset of eliciting contact hypersensitivity in mice: Evidence for a dominant role of TNF-alpha. Journal of Immunology. 1999;162(3):1648–55. [PubMed] [Google Scholar]
- 11.Wang YG, Zhou KJ, Huang G, Hensley C, Huang XN, Ma XP, Zhao T, Sumer BD, DeBerardinis RJ, Gao JM. A nanoparticle-based strategy for the imaging of a broad range of tumours by nonlinear amplification of microenvironment signals. Nat Mater. 2014;13(2):204–12. doi: 10.1038/nmat3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsourkas A, Shinde-Patil VR, Kelly KA, Patel P, Wolley A, Allport JR, Weissleder R. In vivo imaging of activated endothelium using an anti-VCAM-1 magnetooptical probe. Bioconjugate Chem. 2005;16(3):576–81. doi: 10.1021/bc050002e. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, Uertz J, Yohan D, Chithrani BD. Peptide modified gold nanoparticles for improved cellular uptake, nuclear transport, and intracellular retention. Nanoscale. 2014;6(20):12026–33. doi: 10.1039/c4nr02535k. [DOI] [PubMed] [Google Scholar]
- 14.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nature Biotechnology. 1996;14(3):303–8. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nature Biotechnology. 1998;16(1):49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 16.Jayagopal A, Halfpenny KC, Perez JW, Wright DW. Hairpin DNA-Functionalized Gold Colloids for the Imaging of mRNA in Live Cells. Journal of the American Chemical Society. 2010;132(28):9789–96. doi: 10.1021/ja102585v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harry SR, Hicks DJ, Amiri KI, Wright DW. Hairpin DNA coated gold nanoparticles as intracellular mRNA probes for the detection of tyrosinase gene expression in melanoma cells. Chem Commun. 2010;46(30):5557–9. doi: 10.1039/c001969k. [DOI] [PubMed] [Google Scholar]
- 18.Taton TA. Preparation of gold nanoparticle-DNA conjugates. Curr Protoc Nucleic Acid Chem. 2002;Chapter 12(Unit 12):2. doi: 10.1002/0471142700.nc1202s09.. [DOI] [PubMed] [Google Scholar]
- 19.Vesaratchanon S, Nikolov A, Wasan DT. Sedimentation in nano-colloidal dispersions: effects of collective interactions and particle charge. Adv Colloid Interface Sci. 2007;134–135:268–78. doi: 10.1016/j.cis.2007.04.026.. [DOI] [PubMed] [Google Scholar]
- 20.Finkelstein AE, Walz DT, Batista V, Mizraji M, Roisman F, Misher A. Auranofin - New Oral Gold Compound for Treatment of Rheumatoid-Arthritis. Ann Rheum Dis. 1976;35(3):251–7. doi: 10.1136/ard.35.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metz O, Stoll W, Plenert W. Meningosis Prophylaxis with Intrathecal Au-198-Colloid and Methotrexate in Childhood Acute Lymphocytic-Leukemia. Cancer. 1982;49(2):224–8. doi: 10.1002/1097-0142(19820115)49:2<224::Aid-Cncr2820490205>3.0.Co;2-O. [DOI] [PubMed] [Google Scholar]
- 22.Limb GA, Webster L, Soomro H, Janikoun S, Shilling J. Platelet expression of tumour necrosis factor-alpha (TNF-alpha), TNF receptors and intercellular adhesion molecule-1 (ICAM-1) in patients with proliferative diabetic retinopathy. Clin Exp Immunol. 1999;118(2):213–8. doi: 10.1046/j.1365-2249.1999.01067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolle J, Hill RR, Ferguson E, Devall LJ, Trivedi BK, Newton RS, Saxena U. Selective inhibition of tumor necrosis factor-induced vascular cell adhesion molecule-1 gene expression by a novel flavonoid. Lack of effect on transcription factor NF-kappa B. Arterioscler Thromb Vasc Biol. 1996;16(12):1501–8. doi: 10.1161/01.atv.16.12.1501. [DOI] [PubMed] [Google Scholar]
- 24.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129(50):15477–9. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore TL, Rodriguez-Lorenzo L, Hirsch V, Balog S, Urban D, Jud C, Rothen-Rutishauser B, Lattuada M, Petri-Fink A. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chem Soc Rev. 2015;44(17):6287–305. doi: 10.1039/c4cs00487f.. [DOI] [PubMed] [Google Scholar]
- 26.Alkilany AM, Murphy CJ. Toxicity and cellular uptake of gold nanoparticles: what we have learned so far? J Nanopart Res. 2010;12(7):2313–33. doi: 10.1007/s11051-010-9911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du SF, Kendall K, Toloueinia P, Mehrabadi Y, Gupta G, Newton J. Aggregation and adhesion of gold nanoparticles in phosphate buffered saline. Journal of Nanoparticle Research. 2012;14(3) doi: 10.1007/s11051-012-0758-z. UNSP758. [DOI] [Google Scholar]
- 28.Dulkeith E, Ringler M, Klar TA, Feldmann J, Munoz Javier A, Parak WJ. Gold nanoparticles quench fluorescence by phase induced radiative rate suppression. Nano Lett. 2005;5(4):585–9. doi: 10.1021/nl0480969.. [DOI] [PubMed] [Google Scholar]
- 29.Carreon JR, Stewart KM, Mahon KP, Jr, Shin S, Kelley SO. Cyanine dye conjugates as probes for live cell imaging. Bioorg Med Chem Lett. 2007;17(18):5182–5. doi: 10.1016/j.bmcl.2007.06.097.. [DOI] [PubMed] [Google Scholar]
- 30.Miller EW, Zeng L, Domaille DW, Chang CJ. Preparation and use of Coppersensor-1, a synthetic fluorophore for live-cell copper imaging. Nat Protoc. 2006;1(2):824–7. doi: 10.1038/nprot.2006.140.. [DOI] [PubMed] [Google Scholar]
- 31.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312(5776):1027–30. doi: 10.1126/science.1125559.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.