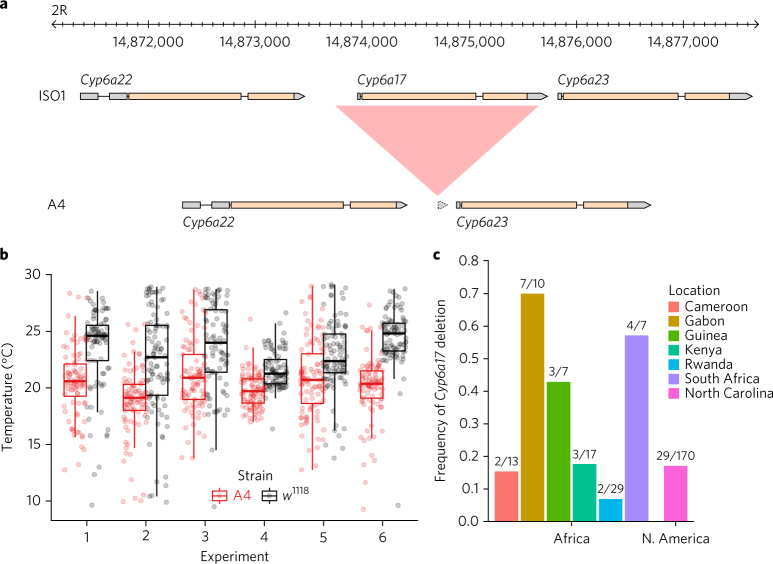
Fig. 2. Copy number variation of Cyp6a17 and its functional consequences.
a, Cyp6a17 is deleted in the A4 genome relative to the ISO1 genome. Alignment between annotated ISO1 and A4 assemblies on chromosome arm 2R shows a large ISO1 region (red) missing in A4. Gene models are shown (gray indicates noncoding sequences, and yellow indicates coding sequences). b, Temperature preference of strains A4 (∆Cyp6a17) and w 1118 (intact Cyp6a17 23). Preference was measured by recording the position of 100 flies along a linear 8 °C–30 °C temperature gradient after an adjustment period (Methods). Each dot represents the position of a fly along the gradient. Each experiment number is an independent pairwise trial. A4 flies occupy colder regions of the gradient than w 1118 flies (Fisher’s method on Wilcoxson rank-sum tests, meta P value << 10−16). Upper and lower hinges of the box plots represent 25% and 75% quantiles, respectively; the upper whisker indicates the largest observation less than or equal to the upper hinge + 1.5 times the interquartile range (IQR); the lower whisker indicates the smallest observation greater than or equal to the lower hinge – 1.5 times the IQR; and the middle horizontal bar indicates the median, 50% quantile. c, Frequency of the Cyp6a17 deletion in African (DPGP2) and North American (DGRP) populations.