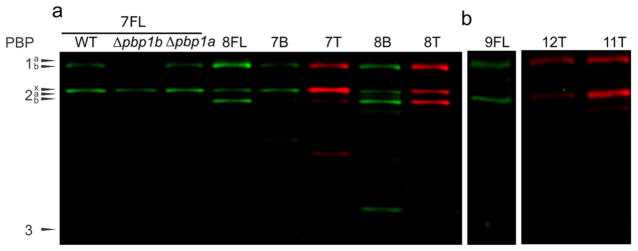
Figure 4.
Gel-based analysis of probes containing side chains to mimic β-lactam antibiotics. (a) L-Phe-(2R,3S)-β-lactone (7T, B, and FL), D-Phe-(2R,3S)-β-lactone (8T, B, and FL) labeling was generally identical between FL and T variants, while BODIPY often increased the number of proteins labeled, including non-PBPs (see 8B). L-Phe-(2R,3S)-β-lactone-based probes (7) showed promise for assessment of PBP2x by use of the Δpbp1b strain. Gel-based analysis of S. pneumoniae wild-type (IU1945), Δpbp1a (E177), and Δpbp1b (E193) strains was performed to confirm specific labeling of PBP2x in the mutant strain. (b) Additional hydrophobic side chains were examined including L-Tyr (9FL), L-Val (11T), and L-Trp (12T), all of which labeled PBP1b and PBP2x. All probes were assessed at 5 μg/mL.