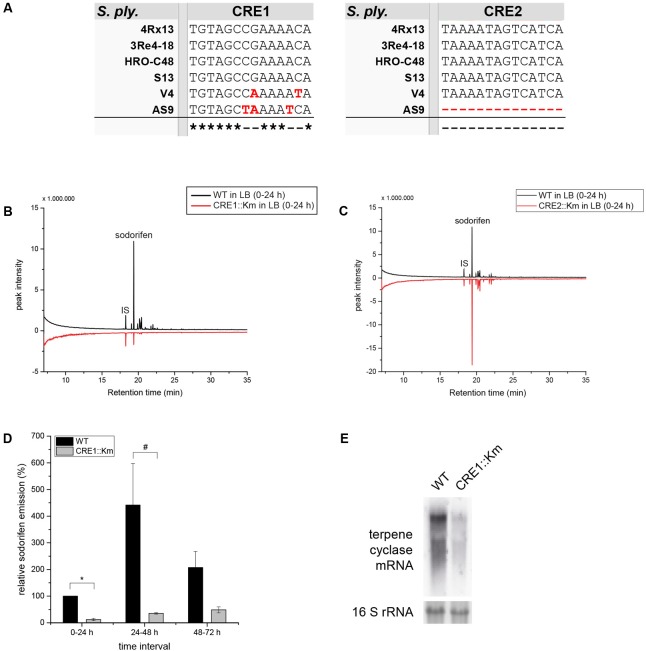
FIGURE 5.
Characterization of CRE1::Km and CRE2::Km deletion mutants. (A) Alignment of the two potential CRE binding sites in the sodorifen cluster upstream sequence from S.p. 4Rx13. Alignment was performed using the Clustal Omega online software (Sievers et al., 2011). Black letters represent nucleotides matching the sequence of S.p. 4Rx13, red letters indicate differences. Asterisks mark identities between all strains tested, dashes represent alterations. (B,C) Sodorifen emission of CRE1::Km/CRE2::Km deletion strains after 24 h cultivation in minimal medium + 55 mM succinate in comparison to the wild type of S.p. 4Rx13. (D) Relative sodorifen emission of CRE1::Km during cultivation in MM + succinate. For calculation of the relative sodorifen emission, the living cell number (CFU/ml) was correlated to the sodorifen amount measured. As a reference, wild type emission until 24 h cultivation was used (100%). Error bars indicate standard deviation (n = 3). ∗p < 0.01; #p < 0.05. Sodorifen emission in (B–D) was determined using the closed VOC collection system (modified after Kai et al., 2010) with subsequent GC/MS analysis. IS, internal standard (nonylacetate, 5 ng). (E) Expression of the terpene cyclase in S.p. 4Rx13 wild type and CRE1::Km. Northern blot was performed with 5 μg RNA isolated after 24 h cultivation in MM + 55 mM succinate. Detection of 16S rRNA expression level was used as a positive control and to ensure equal RNA loading. Expression was detected using DIG – dUTP – labeled probes and fluorescence measurement s for 1 min.