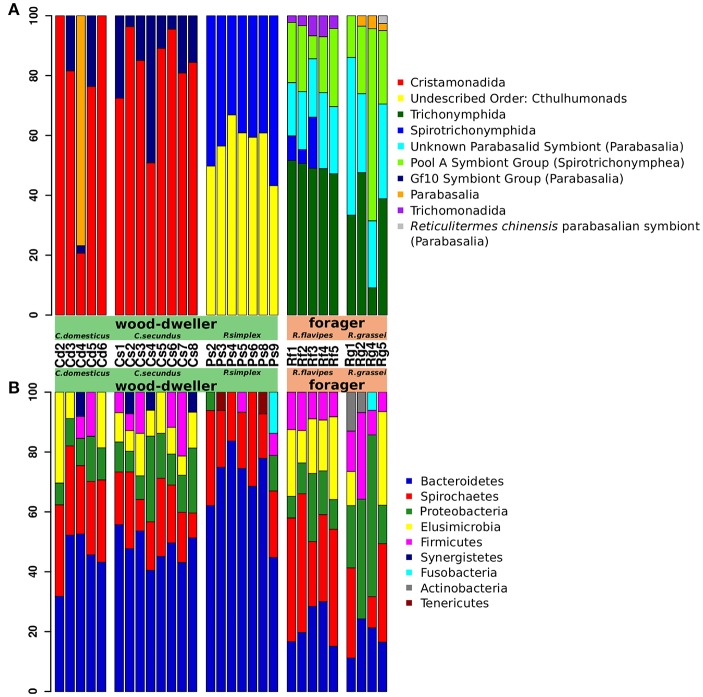
Figure 3.
Termite gut microbiome community composition. Relative abundance of protist and bacterial taxa was assessed using taxonomic profiling of 18S rRNA and 16S rRNA gene amplicon sequences. Taxa are sorted by abundance. (A) Protist community profiles at the order level. Sequences that could not be classified to order with sufficient bootstrap support were labeled with the next higher taxonomic rank with sufficient support. Only orders with an abundance of >2% are shown. Taxa with unconventional names (e.g., “Gf10 Symbiont group”) are followed by the next higher classification in parentheses. “Unknown Parabasalid Symbiont” refers to reference sequences of symbionts isolated from Reticulitermes speratus with yet unidentified taxonomy (Ohkuma et al., 2000). Sequences of the unidentified “Gf 10 symbiont group” were first isolated from Glyptotermes fuscus (Ohkuma et al., 2000) and were also found in other Kalotermitids (Strassert et al., 2009). Note that the taxon “Reticulitermes chinensis parabasalian symbiont” was introduced here for clarification. Both are misleadingly classified as “Cryptotermes symbiont group” in the SILVA taxonomy, although being isolated from Rhinotermitid hosts (SILVA JX975351: isolated from Heterotermes tenuis (James et al., 2013); SILVA GU461593: isolated from R. chinensis, unpublished). (B) Bacterial community composition profiles at the phylum level. Only phyla with a relative abundance of >5% are shown. Cd, colonies of C. domesticus; Cs, C. secundus; Ps, P. simplex; Rf, R. flavipes; Rg, R. grassei.