FIGURE 4.
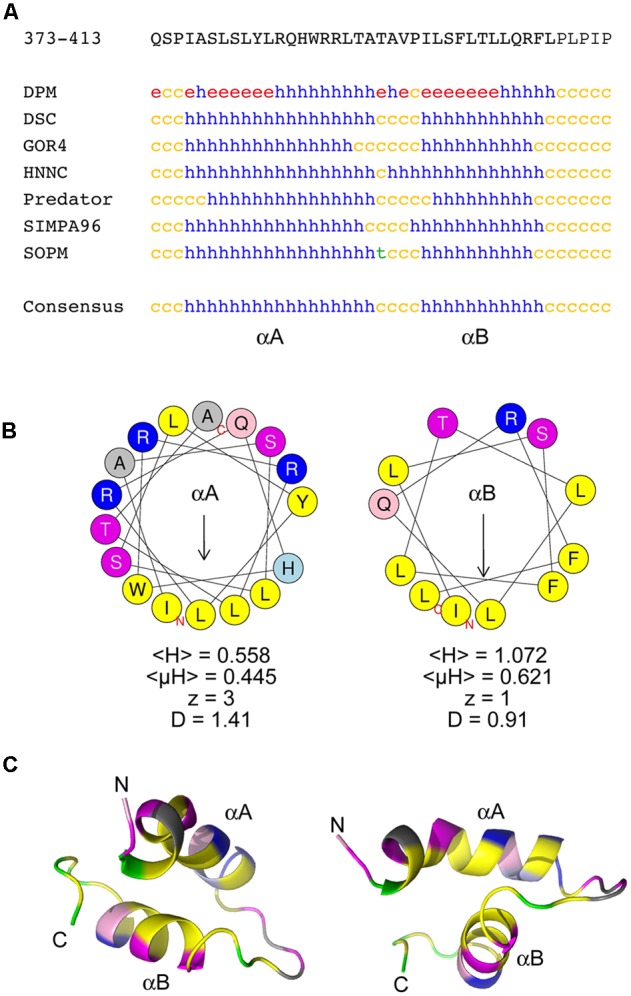
The chloroplast targeting domain contains two predicted amphipathic alpha-helices. (A) Sequence and secondary structure predictions of the CTD domain within the 140K/98K protein. The sequence in one-letter code is shown at the top. Secondary structure predictions were made using several predictors from the NPS@ server (Combet et al., 2000). h: helix; c: coil; e: β-sheet; t: turn). The consensus prediction shown below identifies two α-helices designated αA and αB. (B) Helical wheel representation of αA and αB helices generated using the HeliQuest server (Gautier et al., 2008), illustrating the strong amphipathic character of both αA and αB. Yellow: hydrophobic residues; purple: serine and threonine residues; dark blue: basic residues; light blue: histidine residues; pink: glutamine residues; gray: other residues. The position of the first (N) and last (C) amino acids of the corresponding peptide sequences are indicated. For each helix, the mean hydrophobicity <H>, the mean hydrophobic moment <μH> (in arbitrary units), the charge z and the discriminant factor D are also indicated. The length of the arrow is proportional to the mean hydrophobic moment <μH>. (C) Ab initio modeling of the CTD using PEP-FOLD3 (Lamiable et al., 2016). The top model output is displayed as ribbons and colored with the same color code as in (B), with proline residues in green. The position of the first (N) and last (C) amino acids of the corresponding peptide sequences are indicated. Two different views are shown to illustrate the importance of the linker sequence in the orientation of the helices relative to each other.
