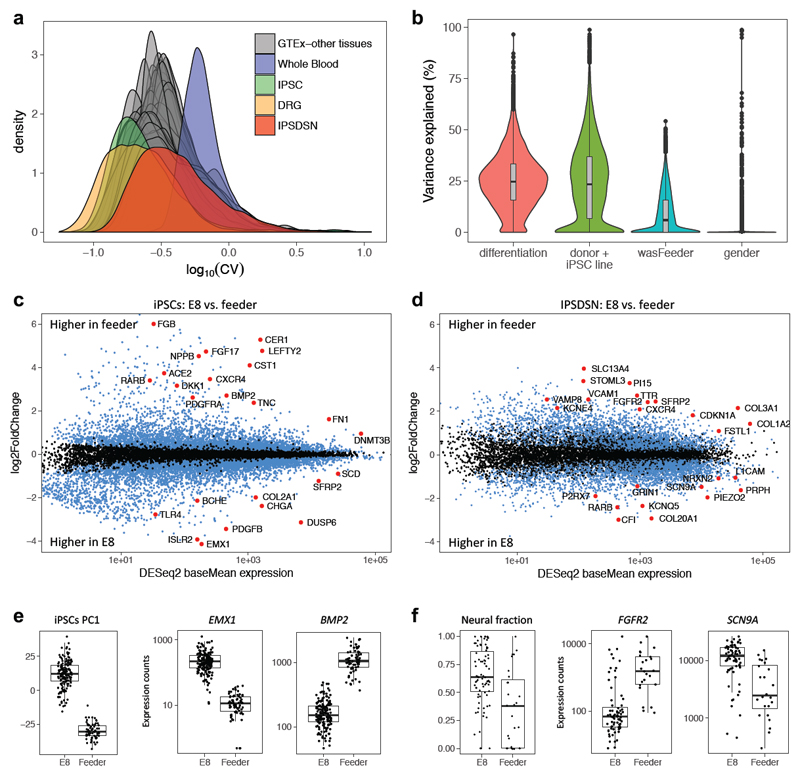
Figure 3. Gene expression variability in IPSDSNs is influenced by differentiation conditions.
(a) Density plot of the coefficient of variation of genes across samples, separately for each GTEx tissue, IPSDSN samples (n=106, P2 protocol only), iPSC (n=239), and DRG (n=28). (b) Violin plot showing, for each gene, the estimated fraction of total expression variability across samples due to differentiation batch, donor genetics or iPSC reprogramming, culture conditions (“wasFeeder”: feeder-dependent vs. E8 medium), and gender. (c) Differentially expressed genes (FDR 1%, blue and red points) between iPSC samples grown on feeders (n=68) vs. E8 medium (n=171). (d) Differentially expressed genes (FDR 1%) between IPSDSNs from feeder- (n=27) and E8-iPSCs (n=79). Neuronal differentiation genes, such as RET and L1CAM, are more highly expressed in samples from E8-iPSCs. (e) Left boxplot: global gene expression differences between feeder- and E8-iPSCs are captured in PC1. Right two boxplots: selected differentially expressed genes. (f) Left boxplot: estimated neural fraction of samples differs in IPSDSNs derived from feeder- and E8-iPSCs. Right two boxplots: selected differentially expressed genes. Boxplots show the median, 25th and 75th percentiles, with whiskers extending 1.5 times the interquartile range.