Figure 4.
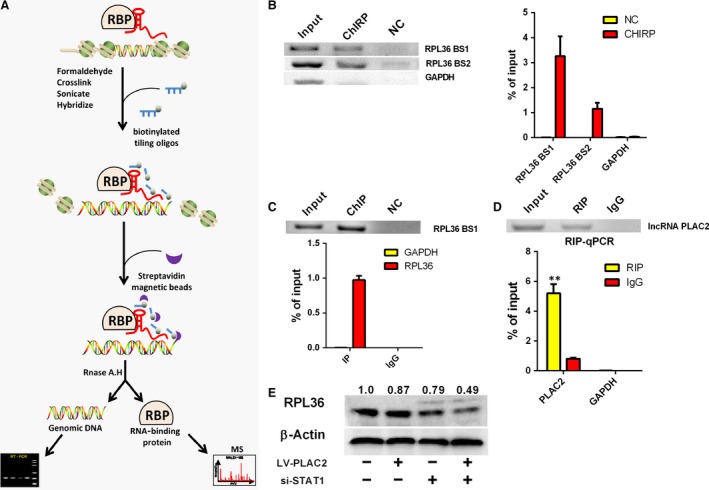
PLAC2 interacts with the transcription factor STAT1. (A) Workflow of ChIRP. Chromatin was cross‐linked to lncRNA:protein adducts in vivo. Biotinylated tiling probes were hybridized to the target lncRNA, and chromatin complexes were purified using magnetic streptavidin beads, followed by stringent washes. (B) Left: Detection of RPL36 DNA in the extracted DNA fraction of ChIRP samples by northern blotting; GAPDH DNA was not detected in the extract; Right: qRT‐PCR analysis revealed an enrichment of RPL36 DNA in the extracted DNA fraction of ChIRP samples. RPL36 DNA was amplified using two promoter‐specific forward and reverse primer pairs, RPL36 BS1 and RPL36 BS2. (C) U87MG cell lysates were subjected to ChIP using an anti‐STAT1 antibody. Top: Northern blot analysis of RPL36 DNA expression. Below: qRT‐PCR analysis of RPL36 DNA expression; >97% of RPL36 DNA was pulled down from the cell by hIP, while no GAPDH was detected. (D) Top: Detection of PLAC2 in the extracted RNA fraction of RIP samples, as detected by Northern blotting. Below: qRT‐PCR analysis of PLAC2 in the extracted RNA fraction of RIP samples, **P < 0.01. (E) STAT1 knockdown in PLAC2‐overexpressing cells enhanced the decrease in RPL36 level in U87MG cells. β‐Actin was used as a loading control.
