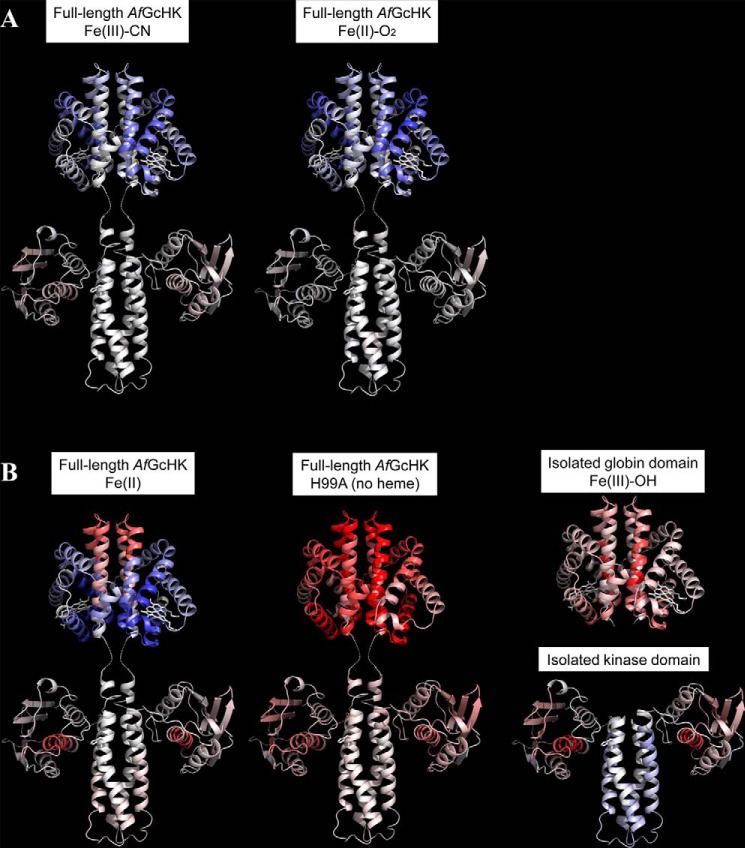
Figure 5.
Conformational changes revealed by HDX–MS after 60 min of deuteration of the full-length AfGcHK proteins and the separated domains, visualized on the protein's structure. Differences between the Fe(III)-OH− form and other active or inactive forms are color-coded; gray indicates no difference, whereas higher and lower levels of deuteration in the specified protein form are indicated by increasingly intense red and blue coloration, respectively. A, active full-length AfGcHK proteins with Fe(III)-CN− (left) and Fe(II)-O2 (center) heme complexes. B, inactive structures, including the full-length AfGcHK protein with an Fe(II) heme complex (left), the full-length heme-free (apo) H99A mutant (center), the isolated globin domain (top right), and the isolated kinase domain (bottom right). The globin domain structure is based on the X-ray crystal data (PDB code 5OHE) presented in this work, and the kinase domain was modeled as an asymmetric homodimer using subunits A and C of the sensor histidine kinase (HK853) from Thermotoga maritima (PDB code 3DGE). This approach was validated by experiments presented elsewhere (7).