Abstract
“Precision Medicine” embodies the analyses of extensive data collected from patients and their environments to identify and apply patient-specific prophylactic strategies and medical treatments to improve clinical outcomes and healthcare cost-effectiveness. Many new methods have been developed for evaluating the activity of the human immune system. Such “immune monitoring” approaches are now being used in studies of allergy and asthma in the hope of identifying better correlates of disease status, predictors of therapeutic outcomes, and potential side-effects of treatment. Together with analyses of family histories, genetic and other biometric data, and measurements of exposures to environmental and other risk factors for developing or exacerbating disease, immune monitoring approaches promise to enable “Precision Medicine” for allergic diseases and asthma.
Keywords: Allergy, asthma, atopic dermatitis, exposome, gene-environment interactions, immune monitoring, metabolome, microbiome, personalized medicine, pharmacogenomics
Precision Medicine and Immune Monitoring
Concepts of “Precision Medicine” (i.e., the patient-specific tailoring of medical treatment based on detailed phenotyping and characterization of the patient, their disease, and their environment) have long been applied in Clinical Allergy/Allergology. In their seminal description of therapeutic immunization with grass pollen extract of subjects suffering from grass pollen allergic rhinitis, Noon and Freeman recognized that the allergen therapy needed to match the cause of the patient’s disease[1–3]. Accordingly, as recently reviewed[4], a foundation of “Precision Medicine” in modern allergology is the initial identification of the patient’s allergic sensitivities prior to forming a plan of treatment, and the assessment of how those sensitivities may be altered by immunotherapy (IT).
The development of genome sequencing, as well as microbiome and virome characterization, stemming from improvements in DNA sequencing methods, has opened the possibility of collecting unprecedentedly detailed genetic data from allergic or asthmatic patients and their commensal or pathogenic microbes. Other ‘omic’ analyses that can be applied to the analysis of allergic disorders and asthma include measurement of ‘epigenetic’ chemical modifications of a person’s genome, characterization of proteins and other macromolecules in bodily fluids or tissues, and measurement of glycan modifications of proteins. Similarly, characterization of the patient’s ‘exposome’ (i.e., the carefully documented record of exposure to components of the patient’s environment, including allergenic plants, animals, fungi, and foods, as well as to therapeutics, tobacco, pollutants, and irradiation) should be considered critical for understanding allergic or asthmatic patients in the context of Precision Medicine (Table 1).
Table 1.
General principles of precision medicine for allergic disorders and asthma*
| Characterize the disease: Identify the disease and, if applicable, the subtype of allergic disorder or asthma; for allergic disorders, precisely define the offending allergen proteins. |
| Profile the patient: Characterize patient genotype and phenotype (and in some cases microbiomes) and their environment (i.e., their “exposome”); assess patient’s likelihood to respond to pharmacological or biological agents, AIT or other forms of management. |
| Select optimal management: Based on the individual’s subtype of disease, offending allergens, genetic and phenotypic characteristics, and an evidence-based assessment of her/his likelihood to respond to various treatment/management options. |
| Monitor disease and response to management: Perform appropriate biometric monitoring during treatment (e.g., with pharmacological or biological agents or with AIT) to assess favorable or adverse effects of the intervention and duration of favorable effects. |
| Develop algorithms to select the most cost-effective management approach for that patient: Based on the characteristics of the patient and his or her test results and the evidence-based assessment of the clinical utility of the treatment options and the type of health care system in which that patient receives his or her care. |
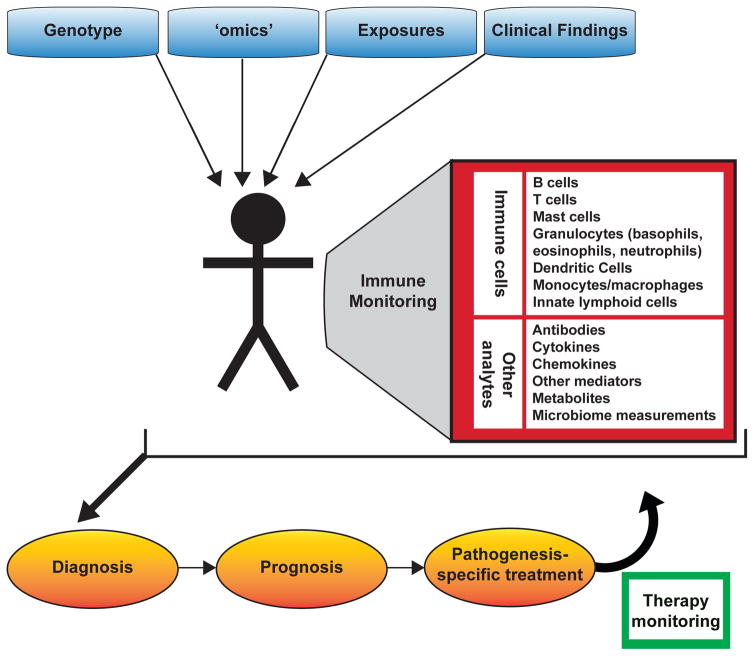
By taking advantage of ongoing basic, translational and clinical research, and having access to patient-specific data obtained during the course of clinical care, these approaches can be continuously and iteratively refined and improved (see Fig 1). This is a modified version of Table 1 in [4], reproduced with permission of the American Academy of Allergy, Asthma, and Immunology.
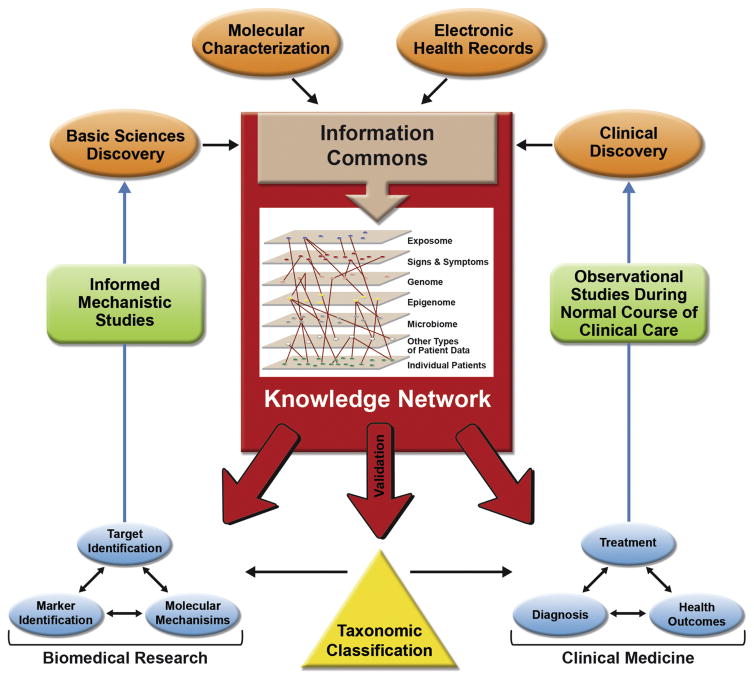
A report from the U.S.A. National Research Council[5] proposed a framework for making sense of large datasets from patients to guide Precision Medicine therapeutic interventions, and to revise disease classifications based on new data gained through such efforts (Fig. 1). The traditional randomized clinical trial[4,5] will still remain the gold standard for assessing the safety and efficacy of any medical intervention. However, recent studies suggest that improved immune monitoring assays may be able to improve our ability to diagnose and evaluate immunological diseases, and predict therapeutic outcomes and side effects. In this review, we will provide some recent examples of work in this broad area.
Figure 1.
Allergic Disease and Asthma Diagnosis and Patient Stratification
Efforts to identify patients with “subtypes” of allergic disorders involve both defining the observable characteristics of their disease (i.e., the disease “phenotype”) and attempting to determine the underlying biological mechanisms involved in the origins and manifestations of the disease (i.e., the disease “endotype”). For example, increasing understanding of the mechanisms underlying food allergy[6–10] is helping to improve diagnosis and stratification of patients[11]. Notably, while allergen-specific serum immunoglobulin E (IgE) and skin prick tests (SPTs) can assist in food allergy diagnoses, these assays are not perfectly sensitive or specific[12]. The more expensive, laborious and higher-risk double-blind, placebo-controlled food challenge (DBPCFC), in which food allergens are administered to patients under carefully monitored conditions to detect clinical reactivity, is the diagnostic gold standard[13]. Alternative tests that do not involve triggering an allergic reaction would represent a significant advance. The use of recombinant allergens[14], and approaches for identifying the allergen epitopes recognized by a patient’s IgE[15–17], may accelerate improvements in diagnostic specificity.
Other promising non-invasive immune monitoring tests include basophil activation tests (BATs), which measure blood basophil activation upon allergen challenge[18,19]. Blood basophil phenotype and function in vitro may be useful in distinguishing between peanut-sensitized children who have clinical allergy versus those who are sensitized but tolerant to peanut[19]. Moreover, it recently was shown that both conventional BATs (i.e., assessment of surface levels of CD63 or CD203c) and cytometry by time-of-flight mass spectrometry (CyTOF) analysis of basophils are robust assays that can be applied to samples shipped overnight, promising improved standardization of such testing at specialized reference laboratories[20]. The application of CyTOF technology[21] to the analysis of basophils[20,22] may reveal previously unsuspected heterogeneity in these cells, and provide additional diagnostic, prognostic and therapeutically-relevant data. Further, two recent reports of new fluorescent-avidin-based assays for basophil activation in whole blood describe a fast and inexpensive BAT that could reveal basophil heterogeneity between different individuals at diagnosis or during therapy[23,24].
In the asthma field, diagnostic classification is evolving rapidly[25], and several new subcategories or “endotypes” of asthma (i.e., variants of asthma that appear to differ in underlying biological mechanisms) were recently reported[26–31], encompassing different genetic variants, patterns of gene expression, and clinical phenotypic features. Efforts to further refine the classification of clinically important subtypes of asthma now include large clinical trial groups in several countries. This work should clarify the requirement for allergen-specific IgE in the pathogenesis of asthma endotypes from early life onward[32–34]. The proposed asthma endotype categories often rely on identifying common downstream pathways (e.g., the detection of a “TH2 cell signature”[31,35]). These downstream pathways are, in some cases, susceptible to targeting with particular biologic therapies[36].
Therapy Selection and Monitoring
Diagnostic methods and classification schemes are of greatest value if they guide selection of effective therapies, and decrease rates of serious side effects. Even if there are reproducible biological differences between subcategories of a disease, unless there are meaningful therapeutic options for each subcategory, or other advantages such as improved prognostication of the disease course, the clinical relevance of the classification will be questionable[4,37,38]. Validation of new classification schemes via testing of different therapies, such as monoclonal antibody drugs, will have major clinical and financial consequences for patients, and for companies developing novel therapies and “companion diagnostics” intended to evaluate whether a therapy would be effective in an individual patient.
In asthma, there are several examples of therapeutic selection based in part on characterization of patient immune responses. Improved identification of patients whose disease has a “TH2 endotype”[31,35], using biomarkers including periostin[39,40] and high levels of blood eosinophils[41], has been used to recommend therapies targeting TH2 response components (e.g., IL-4, IL-5, IL-13 and/or their receptors)[42–47]. However, it appears that there are many pitfalls in attempts to develop these new diagnostic categories for patient populations differing by age, race, or other factors that may alter the correlation of disease phenotype with individual biomarkers. For example, blood periostin levels may not be a useful biomarker of asthma in pediatric populations[48,49], while biomarkers such as exhaled NO and blood eosinophil counts may better predict asthma morbidity in this population[49]. As another example, therapies targeting eosinophils might be most helpful in patients with elevated levels of blood or tissue eosinophils[41,50–54].
Of course, immune monitoring approaches need to be evaluated in clinical trials to determine their merit. A recent study[55] of over 1,000 patients found that optimal “asthma control signatures” identified in whole peripheral blood specimens were enriched for immature lymphocytic gene expression patterns, and that suboptimal control was associated with signatures of eosinophilic and granulocytic inflammation, suggesting that such transcriptional data could guide treatment choices. However, we agree with Gomez and Kaminski[56] in thinking that large prospective clinical studies will be needed to test the usefulness of these immune signatures in predicting suboptimal asthma control over time. It may be possible to refine such approaches by identifying specific cells and regulatory events underlying such immune signatures, both in asthma and other settings[57–60]. The need for large prospective studies to evaluate the clinical utility of immune monitoring assays is similar to proposals for validating pharmacogenomic assays and predictions, including in the area of asthma therapy[61,62].
Similar points can be made regarding identifying any proposed biomarkers for allergic disorders and asthma, as described in recent papers on the potential use of immune monitoring and other data to improve the classification and management of allergic disease, including allergic rhinoconjunctivitis[63] and asthma[36]. As noted by Muraro et al., the heterogeneity of asthma, rhinitis, and AD biomarkers, and variation in the onset, clinical presentation and rates of remission or progression in these diseases combine to generate difficulties in determining the appropriate clinical management strategies, and in selecting biomarkers of therapeutic efficacy[36]. For example, a recent paper suggests that the tyrosine kinase inhibitor, imatinib, may have utility in the treatment of certain patients with severe asthma, but it is unclear whether the critical target is mast cells or other KIT positive cells, or may be due in part to effects of the drug on other tyrosine kinases[64,65]. Perhaps the development of immune profiling tools for assessing the importance of mast cells in asthma could be used to refine the selection of patients for this targeted treatment. That study, which included measurements of the mast cell product, tryptase, in the blood, as well as bronchial biopsies to quantify mast cell numbers in the airways, raises a general point about the extent to which measurements of cell populations and other analytes in the blood can be useful in efforts to “monitor” immune responses whose clinical manifestations reflect the pathology which they induce at specific sites of disease (e.g., the lungs, GI system, or skin). This is an important general problem for the field, and one that is not trivial to study.
Improved immune monitoring approaches could also be relevant in efforts to prevent the development of allergic disease. For example, it is known that genetic predisposition, and the phenotypic manifestation of clinical atopy, are correlated with the development of allergies, including food allergies[66–68]. The recently published LEAP (Learning Early About Peanut) randomized clinical trial provided convincing evidence that children in an intention-to-treat population who had atopy (and therefore had a high probability of developing clinical allergy to peanuts), but who had negative results in SPTs for peanut, had an approximately 85% reduction in development of peanut allergy by 5 years of age if peanut products were introduced into their diet within their first year of life, in contrast to infants avoiding peanuts[69]. This surprising result has led to a reversal of decades of pediatrician advice that parents should avoid exposing their infants to allergenic foods until later in childhood. It may be the case that additional phenotyping of infants, based on the collection of immunological and other clinical data, family histories, and genetic data, could provide the basis for further improvements in decreasing the risk of developing severe allergies by guiding the timing and extent of exposures to environmental factors such as foods associated with allergies. In principle, immune monitoring tests could be less expensive and invasive alternatives to DBPCFC studies for assessing the efficacy of these interventions. Similarly, such assays could be used to evaluate factors that can influence a patient’s threshold for developing a clinical reaction to allergen exposure, such as exercise, alcohol consumption, and concurrent infection[70–72].
A critically important area for implementing Precision Medicine concepts in allergy treatment is in the improvement of Allergen-Specific Immunotherapy (AIT) protocols. Ideally, detailed patient phenotyping could help tailor the allergen dose escalation schedule for AIT for allergic rhinitis[73–75] or oral immunotherapy (OIT) for food allergies, and predict which patients will become permanently desensitized or tolerized[76–78] rather than achieving desensitization dependent on continuing exposure to the allergen[74]. Recent studies provide proof-of-concept data indicating that certain biological measurements can better classify patients and guide their AIT regimens. A recent small phase I single-center clinical trial of OIT for peanut identified FOXP3 gene methylation levels in regulatory T cells as a correlate in patients who achieved more sustained unresponsiveness to peanut after a period without peanut ingestion[77]. Another recent paper based on a small number of subjects identified expanded allergen-specific CD4+ T cells with an “anergic” TH2 T cell phenotype as a feature of patients undergoing OIT[79]. Others have reported that patients undergoing allergen-specific IT show an increased proportion of regulatory T follicular helper T cells to T follicular helper cells[80]. Similarly, analysis of the frequencies of peanut allergen-specific B cells in patients undergoing peanut OIT showed increased levels of specific cells as a correlate of treatment[81,82]. As noted above, recent work indicates that blood basophil phenotype and function analyzed in vitro may be able to distinguish between peanut-sensitized children who have clinical allergy, in contrast to those who are sensitized but tolerant to peanut[19]. Basophil assays also are under investigation for monitoring the efficacy of IT for SAR[83] or omalizumab treatment for severe peanut allergy[84].
Further work with much larger patient cohorts and integrated analyses of different leukocyte populations and other immunological parameters will offer the prospect of identifying the immunological changes that are most closely correlated with the safety and efficacy of treatment, as well as the durability of patient responses. It also will be important to define which of such immune monitoring tests have the greatest clinical utility (and are most cost-effective) for use in routine “check-ups” of patients with allergic diseases, or those at substantial risk to develop such disorders, to enable early detection of allergic diseases, document sustained favorable responses to treatment, and/or give an early indication of the need to consider altering that individual’s management. The results of such efforts, that should include studies of people representing the full diversity of the human population in terms of sex, life stage, genetic background, environmental exposures, and socioeconomic circumstances, will be critical for generating sufficient scientific and clinical data to determine whether a new diagnostic classification of allergic disorders (i.e., a “new taxonomy[5]” of these diseases) should be considered.
Summary
The concepts of Precision Medicine are clearly applicable to the study and classification of allergic disorders and asthma, as well as the selection and monitoring of therapeutic strategies for patients (Table 1, Fig. 2). We feel that the field is only at the beginning of the process of critically evaluating the clinical utility of such “immune monitoring” approaches, some of which involve very newly developed assays. Fundamental questions remain about such approaches, including the extent to which the evaluation of features of immune responses that can be measured in the peripheral blood accurately reflect immunopathology at the tissue sites of disease (Table 2). Ongoing and future work will determine whether immune monitoring approaches will improve disease classification, therapeutic choice, and monitoring of disease status and responses to treatment, as well as the cost-effectiveness of care, in patients with allergic diseases and asthma.
Figure 2.
Table 2.
Needs for advancing precision medicine for allergic disorders and asthma.
| Identify biological factors contributing to development of allergy/asthma |
|
| Identify pathogenic immune system features |
|
| Identify correlates and mechanisms of immunotherapy efficacy |
|
| Identify new therapeutic strategies |
|
| Evaluate cost-effectiveness of new therapeutic strategies |
|
Acknowledgments
We thank members of the Boyd, Nadeau and Galli labs for their contributions to some of the studies reviewed herein, and we thank our colleagues at Stanford, NIAID, and in the allergy research community for many helpful discussions and publications that helped shape our perspectives on these topics. We apologize to the many colleagues whose valuable contributions were not reviewed in this short article, solely due to lack of space.
Funding: This work was supported by the NIH[grant numbers U19AI104209, R01AR067145, and R01AI125567]; the Sean N. Parker Center for Allergy and Asthma, Stanford University, Stanford, CA; and the Department of Pathology, Stanford University, Stanford, CA.
Abbreviations
- AIT
allergen-specific immunotherapy
- BAT
basophil activation test
- CD-sens
basophil allergen threshold sensitivity
- CyTOF
cytometry by time-of-flight mass spectrometry
- DAO
diamine oxidase
- DBPCFC
double-blind, placebo-controlled food challenge (DBPCFC)
- IgE
Immunoglobulin E (antibody)
- IT
immunotherapy
- LEAP
Learning Early About Peanut (a clinical trial)
- OIT
oral immunotherapy
- SAR
seasonal allergic rhinitis
- sIgE
specific Immunoglobulin E (antibody)
- SLIT
sublingual immunotherapy
- SPT
skin prick test
- Tfh
T follicular helper cell
- TH2
T helper cell type 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Freeman J. FURTHER OBSERVATIONS ON THE TREATMENT OF HAY FEVER BY HYPODERMIC INOCULATIONS OF POLLEN VACCINE. The Lancet. 1911;178:814–817. [PubMed] [Google Scholar]
- 2.Freeman J. VACCINATION AGAINST HAY FEVER: REPORT OF RESULTS DURING THE LAST THREE YEARS. The Lancet. 1914;183:1178–1180. [Google Scholar]
- 3.Noon L. PROPHYLACTIC INOCULATION AGAINST HAY FEVER. The Lancet. 177:1572–1573. [Google Scholar]
- 4.Galli SJ. Toward precision medicine and health: Opportunities and challenges in allergic diseases. J Allergy Clin Immunol. 2016;137:1289–1300. doi: 10.1016/j.jaci.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desmond-Helmann SSC, Cox DR, Fraser-Liggett C, Galli SJ, Goldstein DB, et al. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington DC: National Research Council of the National Academies. The National Academies Press; 2011. [PubMed] [Google Scholar]
- 6.Berin MC, Sampson HA. Food allergy: an enigmatic epidemic. Trends Immunol. 2013;34:390–397. doi: 10.1016/j.it.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berin MC, Sicherer S. Food allergy: mechanisms and therapeutics. Curr Opin Immunol. 2011;2:794–800.3. doi: 10.1016/j.coi.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Chinthrajah RS, Hernandez JD, Boyd SD, Galli SJ, Nadeau KC. Molecular and cellular mechanisms of food allergy and food tolerance. J Allergy Clin Immunol. 2016;137:984–997. doi: 10.1016/j.jaci.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wawrzyniak P, Akdis CA, Finkelman FD, Rothenberg ME. Advances and highlights in mechanisms of allergic disease in 2015. J Allergy Clin Immunol. 2016;137:1681–1696. doi: 10.1016/j.jaci.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Yu W, Freeland DM, Nadeau KC. Food allergy: immune mechanisms, diagnosis and immunotherapy. Nat Rev Immunol. 2016;16:751–765. doi: 10.1038/nri.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Muraro A, Lemanske RF, Jr, Castells M, Torres MJ, Khan D, Simon H-U, Bindslev-Jensen C, Burks W, Poulsen LK, Sampson HA, et al. Precision medicine in allergic disease—food allergy, drug allergy, and anaphylaxis—PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy. Asthma and Immunology Allergy. 2017;72:1006–1021. doi: 10.1111/all.13132. This comprehensive consensus document summarizes current thoughts about the potential for precision medicine in food allergy, drug allergy, and anaphylaxis under the auspices of the PRACTALL collaboration platform, which is attempting to harmonize European and American approaches to allergy care. The authors note that although substantial progress has been made in defining endotypes for asthma, such efforts have not made as much progress in food and drug allergy or for anaphylaxis. Some progress has been made in discovery of biomarkers to guide precision medicine approaches for treatment of food and drug allergy, but further validation of these biomarkers is needed to warrant their translation into clinical practice. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton RG. Allergic sensitization is a key risk factor for but not synonymous with allergic disease. J Allergy Clin Immunol. 2014;134:360–361. doi: 10.1016/j.jaci.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Bock SA, Sampson HA, Atkins FM, Zeiger RS, Lehrer S, Sachs M, Bush RK, Metcalfe DD. Double-blind, placebo-controlled food challenge (DBPCFC) as an office procedure: a manual. J Allergy Clin Immunol. 1988;82:986–997. doi: 10.1016/0091-6749(88)90135-2. [DOI] [PubMed] [Google Scholar]
- 14.Mothes N, Valenta R, Spitzauer S. Allergy testing: the role of recombinant allergens. Clin Chem Lab Med. 2006;44:125–132. doi: 10.1515/CCLM.2006.024. [DOI] [PubMed] [Google Scholar]
- 15.Lupinek C, Wollmann E, Baar A, Banerjee S, Breiteneder H, Broecker BM, Bublin M, Curin M, Flicker S, Garmatiuk T, et al. Advances in allergen-microarray technology for diagnosis and monitoring of allergy: the MeDALL allergen-chip. Methods. 2014;66:106–119. doi: 10.1016/j.ymeth.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treudler R, Simon JC. Overview of component resolved diagnostics. Curr Allergy Asthma Rep. 2013;13:110–117. doi: 10.1007/s11882-012-0318-8. [DOI] [PubMed] [Google Scholar]
- 17.Tuano KS, Davis CM. Utility of Component-Resolved Diagnostics in Food Allergy. Curr Allergy Asthma Rep. 2015;15:32. doi: 10.1007/s11882-015-0534-0. [DOI] [PubMed] [Google Scholar]
- 18•.Hoffmann HJ, Knol EF, Ferrer M, Mayorga L, Sabato V, Santos AF, Eberlein B, Nopp A, MacGlashan D. Pros and Cons of Clinical Basophil Testing (BAT) Curr Allergy Asthma Rep. 2016;16:56. doi: 10.1007/s11882-016-0633-6. This is a comprehensive review, which includes an annotated bibliography, of the advantages and disadvantages of basophil testing by flow cytometry. The authors review several studies that have addressed the technical aspects of various methods and their potential clinical relevance for disease diagnosis and immune monitoring. [DOI] [PubMed] [Google Scholar]
- 19.Santos AF, Douiri A, Becares N, Wu SY, Stephens A, Radulovic S, Chan SM, Fox AT, Du Toit G, Turcanu V, et al. Basophil activation test discriminates between allergy and tolerance in peanut-sensitized children. J Allergy Clin Immunol. 2014;134:645–652. doi: 10.1016/j.jaci.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Mukai K, Gaudenzio N, Gupta S, Vivanco N, Bendall SC, Maecker HT, Chinthrajah RS, Tsai M, Nadeau KC, Galli SJ. Assessing basophil activation by using flow cytometry and mass cytometry in blood stored 24 hours before analysis. J Allergy Clin Immunol. 2017;139:889–899. e811. doi: 10.1016/j.jaci.2016.04.060. In an analysis utilizing blood from 46 healthy blood donors and 120 patients with peanut allergy, Mukai et al. showed that both conventional BATs (upregulation of CD63 and CD203c) and CyTOF analysis of basophils can be performed on blood that is anticoagulated with heparin and stored at 4°C for 24 hours. This may allow improved standardization of such testing, e.g., at specialized reference laboratories. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Tordesillas L, Rahman AH, Hartmann BM, Sampson HA, Berin MC. Mass cytometry profiling the response of basophils and the complete peripheral blood compartment to peanut. J Allergy Clin Immunol. 2016;138:1741–1744. e1749. doi: 10.1016/j.jaci.2016.06.048. This study employed CyTOF to analyze the responses to ex vivo stimulation, with peanut extract, anti-IgE or medium, of all peripheral blood cells in blood from 6 peanut allergic patients and 3 healthy controls. In addition to providing a detailed CyTOF profile of many types of blood leukocytes, the authors found that basophils and platelets physically interact after peanut allergen exposure, and suggested that such interactions may be relevant during anaphylaxis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joulia R, Mailhol C, Valitutti S, Didier A, Espinosa E. Direct monitoring of basophil degranulation by using avidin-based probes. J Allergy Clin Immunol. 2017 Apr 24; doi: 10.1016/j.jaci.2017.03.030. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 24•.Mukai K, Chinthrajah RS, Nadeau KC, Tsai M, Gaudenzio N, Galli SJ. A new fluorescent-avidin-based method for quantifying basophil activation in whole blood. J Allergy Clin Immunol. 2017 Jun 9; doi: 10.1016/j.jaci.2017.03.052. [Epub ahead of print] These back-to-back studies report a new fluorescent-avidin-based assay for measuring basophil activation in whole blood. This fast and inexpensive BAT can both identify basophils which have been activated ex vivo (e.g., in response to anti-IgE or specific antigen) and also may reveal evidence of ongoing exteriorization of avidin-binding granule contents on the surface of blood basophils at ‘baseline’ (e.g., in subjects with peanut allergy who are enrolled in OIT trials). The assay also may help to reveal basophil phenotypic heterogeneity in the extent of anti-IgE-induced enhanced basophil surface expression of avidin-binding material vs. CD63 among different individuals at diagnosis or during therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, Adcock IM, Bateman ED, Bel EH, Bleecker ER, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 26.Kim TB, Jang AS, Kwon HS, Park JS, Chang YS, Cho SH, Choi BW, Park JW, Nam DH, Yoon HJ, et al. Identification of asthma clusters in two independent Korean adult asthma cohorts. Eur Respir J. 2013;41:1308–1314. doi: 10.1183/09031936.00100811. [DOI] [PubMed] [Google Scholar]
- 27.Lotvall J, Akdis CA, Bacharier LB, Bjermer L, Casale TB, Custovic A, Lemanske RF, Jr, Wardlaw AJ, Wenzel SE, Greenberger PA. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127:355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 28.Simpson JL, Phipps S, Baines KJ, Oreo KM, Gunawardhana L, Gibson PG. Elevated expression of the NLRP3 inflammasome in neutrophilic asthma. Eur Respir J. 2014;43:1067–1076. doi: 10.1183/09031936.00105013. [DOI] [PubMed] [Google Scholar]
- 29.Skloot GS. Asthma phenotypes and endotypes: a personalized approach to treatment. Curr Opin Pulm Med. 2016;22:3–9. doi: 10.1097/MCP.0000000000000225. [DOI] [PubMed] [Google Scholar]
- 30.Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, Chu HW. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med. 1999;160:1001–1008. doi: 10.1164/ajrccm.160.3.9812110. [DOI] [PubMed] [Google Scholar]
- 31.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, Koth LL, Arron JR, Fahy JV. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma and wheezing in the first six years of life. The Group Health Medical Associates. N Engl J Med. 1995;3:133–138.32. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 33.Martinez FD. Gene-environment interactions in asthma: with apologies to William of Ockham. Proc Am Thorac Soc. 2007;4:26–31. doi: 10.1513/pats.200607-144JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sears MR. Predicting asthma outcomes. J Allergy Clin Immunol. 2015;136:829–836. doi: 10.1016/j.jaci.2015.04.048. quiz 837. [DOI] [PubMed] [Google Scholar]
- 35.Agache I, Sugita K, Morita H, Akdis M, Akdis CA. The Complex Type 2 Endotype in Allergy and Asthma: From Laboratory to Bedside. Curr Allergy Asthma Rep. 2015;15:29. doi: 10.1007/s11882-015-0529-x. [DOI] [PubMed] [Google Scholar]
- 36•.Muraro A, Lemanske RF, Jr, Hellings PW, Akdis CA, Bieber T, Casale TB, Jutel M, Ong PY, Poulsen LK, Schmid-Grendelmeier P, et al. Precision medicine in patients with allergic diseases: Airway diseases and atopic dermatitis-PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2016;137:1347–1358. doi: 10.1016/j.jaci.2016.03.010. This thoughtful consensus document summarizes current knowledge of major asthma, rhinitis, and atopic dermatitis endotypes under the auspices of the PRACTALL collaboration platform, which is attempting to harmonize European and American approaches to best allergy practice and science, including opportunities for using precision medicine approaches in the management of asthma, rhinitis, and atopic dermatitis (e.g., in better selection of treatment responders, risk prediction, and design of disease-modifying strategies) [DOI] [PubMed] [Google Scholar]
- 37•.Berry A, Busse WW. Biomarkers in asthmatic patients: Has their time come to direct treatment? J Allergy Clin Immunol. 2016;137:1317–1324. doi: 10.1016/j.jaci.2016.03.009. This review summarizes current efforts to use biomarkers for the diagnosis of asthma, assessing the severity of disease, and evaluating the potential efficacy of treatments. To quote the authors: “Progress has been made in the discovery, application, and implementation of biomarkers in asthmatic patients. The journey is just beginning, and the biomarkers now available, although of considerable benefit, are still too broad, too limited, and too nonspecific to identify who is at risk for asthma, what will be their pattern of disease, and what specific treatments, particularly biologics, will be most helpful. The needs for biomarkers are great and the interest high; the future for their use has yet to be fully achieved.”. [DOI] [PubMed] [Google Scholar]
- 38.Skevaki C, Van den Berg J, Jones N, Garssen J, Vuillermin P, Levin M, Landay A, Renz H, Calder PC, Thornton CA. Immune biomarkers in the spectrum of childhood noncommunicable diseases. J Allergy Clin Immunol. 2016;137:1302–1316. doi: 10.1016/j.jaci.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 39.Izuhara K, Matsumoto H, Ohta S, Ono J, Arima K, Ogawa M. Recent developments regarding periostin in bronchial asthma. Allergol Int. 2015;64(Suppl):S3–10. doi: 10.1016/j.alit.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Matsusaka M, Kabata H, Fukunaga K, Suzuki Y, Masaki K, Mochimaru T, Sakamaki F, Oyamada Y, Inoue T, Oguma T, et al. Phenotype of asthma related with high serum periostin levels. Allergol Int. 2015;64:175–180. doi: 10.1016/j.alit.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Cardet JC, Israel E. Update on reslizumab for eosinophilic asthma. Expert Opin Biol Ther. 2015;1:1531–1539. doi: 10.1517/14712598.2015.1090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, Harris JM, Scheerens H, Wu LC, Su Z, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–1098. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 43.Kau AL, Korenblat PE. Anti-interleukin 4 and 13 for asthma treatment in the era of endotypes. Curr Opin Allergy Clin Immunol. 2014 doi: 10.1097/ACI.0000000000000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noonan M, Korenblat P, Mosesova S, Scheerens H, Arron JR, Zheng Y, Putnam WS, Parsey MV, Bohen SP, Matthews JG. Dose-ranging study of lebrikizumab in asthmatic patients not receiving inhaled steroids. J Allergy Clin Immunol. 2013;132:567–574. e512. doi: 10.1016/j.jaci.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 45.Slager RE, Hawkins GA, Ampleford EJ, Bowden A, Stevens LE, Morton MT, Tomkinson A, Wenzel SE, Longphre M, Bleecker ER, et al. IL-4 receptor alpha polymorphisms are predictors of a pharmacogenetic response to a novel IL-4/IL-13 antagonist. J Allergy Clin Immunol. 2010;126:875–878. doi: 10.1016/j.jaci.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenzel S, Wilbraham D, Fuller R, Getz EB, Longphre M. Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: results of two phase 2a studies. Lancet. 2007;370:1422–1431. doi: 10.1016/S0140-6736(07)61600-6. [DOI] [PubMed] [Google Scholar]
- 47.Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, Wang L, Kirkesseli S, Rocklin R, Bock B, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med. 2013;368:2455–2466. doi: 10.1056/NEJMoa1304048. [DOI] [PubMed] [Google Scholar]
- 48.Inoue Y, Izuhara K, Ohta S, Ono J, Shimojo N. No increase in the serum periostin level is detected in elementary school-age children with allergic diseases. Allergol Int. 2015;64:289–290. doi: 10.1016/j.alit.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Konradsen JR, Skantz E, Nordlund B, Lidegran M, James A, Ono J, Ohta S, Izuhara K, Dahlen SE, Alving K, et al. Predicting asthma morbidity in children using proposed markers of Th2-type inflammation. Pediatr Allergy Immunol. 2015;26:772–779. doi: 10.1111/pai.12457. [DOI] [PubMed] [Google Scholar]
- 50.Castro M, Mathur S, Hargreave F, Boulet LP, Xie F, Young J, Wilkins HJ, Henkel T, Nair P. Reslizumab for poorly controlled, eosinophilic asthma: a randomized, placebo-controlled study. Am J Respir Crit Care Med. 2011;184:1125–1132. doi: 10.1164/rccm.201103-0396OC. [DOI] [PubMed] [Google Scholar]
- 51.Castro M, Wenzel SE, Bleecker ER, Pizzichini E, Kuna P, Busse WW, Gossage DL, Ward CK, Wu Y, Wang B, et al. Benralizumab, an anti-interleukin 5 receptor alpha monoclonal antibody, versus placebo for uncontrolled eosinophilic asthma: a phase 2b randomised dose-ranging study. Lancet Respir Med. 2014;2:879–890. doi: 10.1016/S2213-2600(14)70201-2. [DOI] [PubMed] [Google Scholar]
- 52.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, Hargreave FE, O’Byrne PM. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009;360:985–993. doi: 10.1056/NEJMoa0805435. [DOI] [PubMed] [Google Scholar]
- 54.Nair P. What is an “eosinophilic phenotype” of asthma? J Allergy Clin Immunol. 2013;132:81–83. doi: 10.1016/j.jaci.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 55••.Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF, Jr, Liu AH, Gilliland FD, Millstein J, Gauderman WJ, Ober C, et al. Gene Expression Profiling in Blood Provides Reproducible Molecular Insights into Asthma Control. Am J Respir Crit Care Med. 2017;195:179–188. doi: 10.1164/rccm.201601-0107OC. Using transcriptional profiling of whole blood or CD4+ T lymphocytes from the Asthma BioRepository for Integrative Genomic Exploration and the Childhood Asthma Management Program cohorts, and employing gene set enrichment analyses of asthma control and the immunologic signatures gene set collections of the Molecular Signatures Database, the authors identified molecular pathways associated with a modified version of the asthma control test. They provide evidence that optimal asthma control signatures were enriched for immature lymphocytic gene expression patterns, and that suboptimal control was associated with signatures of eosinophilic and granulocytic inflammation. Notably, their work suggests that the triggering receptor expressed on myeloid cells 1 (TREM1 or CD354, a cell surface receptor expressed on activated monocytes, neutrophils, granulocytes, dendritic cells, and natural killer cells) is involved in asthma control, suggesting that patients with suboptimal asthma control exhibit signs of persistent innate immune activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez JL, Kaminski N. Toward Precision Medicine of Symptom Control in Asthma. Am J Respir Crit Care Med. 2017;195:147–148. doi: 10.1164/rccm.201608-1600ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.Gustafsson M, Gawel DR, Alfredsson L, Baranzini S, Bjorkander J, Blomgran R, Hellberg S, Eklund D, Ernerudh J, Kockum I, et al. A validated gene regulatory network and GWAS identifies early regulators of T cell-associated diseases. Sci Transl Med. 2015;7:313ra178. doi: 10.1126/scitranslmed.aad2722. The authors measured by qPCR splice variants of three transcription factors involved in TH1/TH2 differentiation (GATA3, MAF and MYB) in peripheral blood T cells of patients with several T cell-associated diseases, and used siRNA-mediated knockdown to assess the disease relevance of MAF splice variant 2 in patients with seasonal allergic rhinitis (SAR), following 14 patients before, during (after 1 year), and after 2 years of SLIT. They found that SLIT resulted in reversal of expression of the analyzed splice variants in SAR patients to levels observed in healthy controls, suggesting that this approach might be helpful as a component of immune monitoring in patients undergoing treatment for SAR. [DOI] [PubMed] [Google Scholar]
- 58.Lao-Araya M, Steveling E, Scadding GW, Durham SR, Shamji MH. Seasonal increases in peripheral innate lymphoid type 2 cells are inhibited by subcutaneous grass pollen immunotherapy. J Allergy Clin Immunol. 2014;134:1193–1195. e1194. doi: 10.1016/j.jaci.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 59•.Metcalfe DD, Pawankar R, Ackerman SJ, Akin C, Clayton F, Falcone FH, Gleich GJ, Irani AM, Johansson MW, Klion AD, et al. Biomarkers of the involvement of mast cells, basophils and eosinophils in asthma and allergic diseases. World Allergy Organ J. 2016;9:7. doi: 10.1186/s40413-016-0094-3. This is a concise review of the biomarkers which may be useful in attempting to assess the involvement of mast cells, basophils, and eosinophils in allergic inflammation in humans, including surface markers of cell activation and specific products that implicate these various cell types in allergic diseases and asthma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60•.Rust B, Wambre E. Human Immune Monitoring Techniques during Food Allergen Immunotherapy. Curr Allergy Asthma Rep. 2017;17:22. doi: 10.1007/s11882-017-0689-y. This timely brief review, that includes an annotated bibliography, discusses current technical approaches used in immune monitoring during IT in patients with allergies, particularly focusing on cell-mediated immunity and the analysis of antigen-specific T cells. It also discusses important challenges in this area, including pre-analytical issues and the prerequisites for performing effective immune monitoring. [DOI] [PubMed] [Google Scholar]
- 61•.Kersten ET, Koppelman GH. Pharmacogenetics of asthma: toward precision medicine. Curr Opin Pulm Med. 2017;23:12–20. doi: 10.1097/MCP.0000000000000335. This thoughtful review, which includes an annotated bibliography, notes that many pharmacogenetics findings in asthma have not been replicated and that currently identified genetic loci account for only a small fraction of variability in drug response. They propose that the translation of such pharmacogenetics findings into clinical practice likely will require integration of such data with those derived from the application of other ‘omics’ technologies. They also remind us that direct comparisons of “precision medicine” approaches with traditional clinical management methods may be needed to change clinical practice. [DOI] [PubMed] [Google Scholar]
- 62•.Ortega VE, Meyers DA, Bleecker ER. Asthma pharmacogenetics and the development of genetic profiles for personalized medicine. Pharmgenomics Pers Med. 2015;8:9–22. doi: 10.2147/PGPM.S52846. This is a comprehensive review of pharmacogenetic studies which are helping to define genetic profiles that can be used for personalized medicine in asthma, including candidate gene and genome-wide association studies which have identified genetic loci associated with therapeutic responsiveness in the glucocorticoid, leukotriene, and β2-adrenergic receptor pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Shamji MH, Kappen JH, Akdis M, Jensen-Jarolim E, Knol EF, Kleine-Tebbe J, Bohle B, Chaker AM, Till SJ, Valenta R, et al. Biomarkers for monitoring clinical efficacy of allergen immunotherapy for allergic rhinoconjunctivitis and allergic asthma: an EAACI Position Paper. Allergy. 2017 Feb 2; doi: 10.1111/all.13138. [Epub ahead of print] An EAACI taskforce reviewed the literature regarding all candidate biomarkers used in clinical trials of allergic rhinoconjuctivitis patients with or without asthma: (i) IgE (total IgE, specific IgE [sIgE] and sIgE/total IgE ratio), (ii) IgG-subclasses (sIgG1, sIgG4 including sIgE/IgG4 ratio), (iii) serum inhibitory activity for IgE, (iv) basophil activation, (v) cytokines and chemokines, (vi) cellular markers (T regulatory cells, B regulatory cells and dendritic cells) and (vii) in vivo biomarkers. The authors recommended exploring the use of allergen-specific IgG4 as a biomarker for compliance, as well as more studies to validate other potential biomarkers of clinical responses to allergen-specific IT. [DOI] [PubMed] [Google Scholar]
- 64•.Cahill KN, Katz HR, Cui J, Lai J, Kazani S, Crosby-Thompson A, Garofalo D, Castro M, Jarjour N, DiMango E, et al. KIT Inhibition by Imatinib in Patients with Severe Refractory Asthma. N Engl J Med. 2017;376:1911–1920. doi: 10.1056/NEJMoa1613125. This is the first clinical study to assess the possibility that “targeting” mast cells, in this case with the tyrosine kinase inhibitor, imatinib, might have benefit in the treatment of patients with severe asthma. While the results reported are not clinically directive, they do suggest that the therapeutic targeting of mast cells in certain patients with asthma represents a potentially important topic for future research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galli SJ. Mast Cells and KIT as Potential Therapeutic Targets in Severe Asthma. N Engl J Med. 2017;376:1983–1984. doi: 10.1056/NEJMe1702653. [DOI] [PubMed] [Google Scholar]
- 66.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Portelli MA, Hodge E, Sayers I. Genetic risk factors for the development of allergic disease identified by genome-wide association. Clin Exp Allergy. 2015;45:21–31. doi: 10.1111/cea.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 69••.Du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, Brough HA, Phippard D, Basting M, Feeney M, et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. 2015;372:803–813. doi: 10.1056/NEJMoa1414850. This landmark study, which has been widely discussed in the medical and lay media, reports the results of a randomized clinical trial showing that children in an intention-to-treat population who had atopy, but negative results in SPTs for peanut, had an ~85% reduction in development of peanut allergy by 5 years of age if peanut products were introduced into their diet in their first year of life, in contrast to infants avoiding peanuts. This result led to a reversal of decades of medical advice that parents should avoid exposing their infants to allergenic foods until later in childhood. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hefle SL, Taylor SL. How much food is too much? Threshold doses for allergenic foods. Curr Allergy Asthma Rep. 2002;2:63–66. doi: 10.1007/s11882-002-0041-y. [DOI] [PubMed] [Google Scholar]
- 71.Smith PK, Hourihane JO, Lieberman P. Risk multipliers for severe food anaphylaxis. World Allergy Organ J. 2015;8:30. doi: 10.1186/s40413-015-0081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sicherer SH, Sampson HA. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. 2014;133:291–307. doi: 10.1016/j.jaci.2013.11.020. quiz 308. [DOI] [PubMed] [Google Scholar]
- 73•.Akdis CA, Akdis M. Advances in allergen immunotherapy: aiming for complete tolerance to allergens. Sci Transl Med. 2015;7:280ps286. doi: 10.1126/scitranslmed.aaa7390. This is a thoughtful review of recent advances in the field of allergen-specific immunotherapy, including its potential immunological targets and mechanisms of action, as well as current efforts to improve the standardization and efficacy of such approaches. [DOI] [PubMed] [Google Scholar]
- 74.Frew AJ. Allergen immunotherapy. J Allergy Clin Immunol. 2010;125:S306–313. doi: 10.1016/j.jaci.2009.10.064. [DOI] [PubMed] [Google Scholar]
- 75.Larche M, Akdis CA, Valenta R. Immunological mechanisms of allergen-specific immunotherapy. Nat Rev Immunol. 2006;6:761–771. doi: 10.1038/nri1934. [DOI] [PubMed] [Google Scholar]
- 76.Burks AW, Jones SM, Wood RA, Fleischer DM, Sicherer SH, Lindblad RW, Stablein D, Henning AK, Vickery BP, Liu AH, et al. Oral immunotherapy for treatment of egg allergy in children. N Engl J Med. 2012;367:233–243. doi: 10.1056/NEJMoa1200435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Syed A, Garcia MA, Lyu SC, Bucayu R, Kohli A, Ishida S, Berglund JP, Tsai M, Maecker H, O’Riordan G, et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3) J Allergy Clin Immunol. 2014;133:500–510. doi: 10.1016/j.jaci.2013.12.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vickery BP, Scurlock AM, Kulis M, Steele PH, Kamilaris J, Berglund JP, Burk C, Hiegel A, Carlisle S, Christie L, et al. Sustained unresponsiveness to peanut in subjects who have completed peanut oral immunotherapy. J Allergy Clin Immunol. 2014;133:468–475. doi: 10.1016/j.jaci.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79•.Ryan JF, Hovde R, Glanville J, Lyu SC, Ji X, Gupta S, Tibshirani RJ, Jay DC, Boyd SD, Chinthrajah RS, et al. Successful immunotherapy induces previously unidentified allergen-specific CD4+ T-cell subsets. Proc Natl Acad Sci U S A. 2016;113:E1286–1295. doi: 10.1073/pnas.1520180113. In a study of 5 peanut allergic participants undergoing OIT with peanut and 7 healthy controls, allergen-specific T-cell sorting and single-cell gene expression was used to characterize the transcriptional profile of individual CD4+ T cells during OIT. The authors provide evidence that successful OIT induced allergen-specific CD4+ T cells to expand and shift toward an “anergic” Th2 T-cell phenotype which was largely absent in both pretreatment participants and healthy controls. These findings suggest that sustained success of IT is associated with the induction, expansion, and maintenance of IT-specific memory and naïve T-cell phenotypes as early as 3 months into IT. These results suggest an approach, which would need to be confirmed in larger studies, for immune monitoring of participants undergoing IT to predict the success of future treatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80•.Schulten V, Tripple V, Seumois G, Qian Y, Scheuermann RH, Fu Z, Locci M, Rosales S, Vijayanand P, Sette A, et al. Allergen-specific immunotherapy modulates the balance of circulating Tfh and Tfr cells. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2017.04.032. This study, of 25 timothy grass allergic patients, 32 patients who received subcutaneous shots of allergen-specific IT (AIT) and were in treatment maintenance at the time of blood draw, and 13 nonallergic healthy controls, identified a significant reduction in follicular helper T (Tfh) cells in AIT-treated patients compared with untreated allergic donors (median 6.4% in allergics, 3.5% in AIT-treated patients; P=.008). Based on these and other data, the authors hypothesized that repeated administration of allergen extracts elicits IL-2 production from allergen-specific T-cells, which globally impairs CXCR5 expression in memory Tfh cells and induces/retains Tfr-cell populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81•.Hoh RA, Joshi SA, Liu Y, Wang C, Roskin KM, Lee JY, Pham T, Looney TJ, Jackson KJ, Dixit VP, et al. Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. J Allergy Clin Immunol. 2016;137:157–167. doi: 10.1016/j.jaci.2015.05.029. This paper and the study by Patil et al. below, characterize peanut allergen-specific B cells and sequence their antibody gene rearrangements, in peanut allergic individuals. These represent new steps toward a full molecular definition of the IgE antibodies that are responsible for peanut allergy, as well as peanut-specific non-IgE antibodies that may be involved in therapeutic responses to OIT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82•.Patil SU, Ogunniyi AO, Calatroni A, Tadigotla VR, Ruiter B, Ma A, Moon J, Love JC, Shreffler WG. Peanut oral immunotherapy transiently expands circulating Ara h 2-specific B cells with a homologous repertoire in unrelated subjects. J Allergy Clin Immunol. 2015;136:125–134. e112. doi: 10.1016/j.jaci.2015.03.026. Among the results in this paper and Hoh et al. above, are increases in the frequency of peanut allergen-specific B cells in the blood during OIT, and significant levels of somatic mutation in the allergen-specific antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83•.Shamji MH, Layhadi JA, Scadding GW, Cheung DK, Calderon MA, Turka LA, Phippard D, Durham SR. Basophil expression of diamine oxidase: a novel biomarker of allergen immunotherapy response. J Allergy Clin Immunol. 2015;135:913–921. e919. doi: 10.1016/j.jaci.2014.09.049. This study showed that, in grass pollen allergic individuals with SAR treated with subcutaneous or sublingual IT, intracellular fluorochrome-labeled diamine oxidase (DAO) can be used as functional readout of histamine release by basophils challenged with allergen ex vivo, and that this ex vivo assessment of allergen-induced basophil reactivity correlates with clinical responses to IT. The authors proposed further studies to determine whether intracellularly labeled DAO expression by basophils might be used as a surrogate biomarker for monitoring efficacy of IT and the durability of favorable responses. [DOI] [PubMed] [Google Scholar]
- 84•.Brandström J, Vetander M, Lilja G, Johansson SG, Sundqvist AC, Kalm F, Nilsson C, Nopp A. Individually dosed omalizumab: an effective treatment for severe peanut allergy. Clin Exp Allergy. 2017;47:540–550. doi: 10.1111/cea.12862. In this open phase 2 study of omalizumab treatment in 23 severely peanut allergic patients, measurements of basophil allergen threshold sensitivity (CD-sens) (which correlate with the outcome of DBPCFC with peanut) were used to measure omalizumab treatment efficacy. The authors found that participants who needed an elevated omalizumab dose (ED) to suppress CD-sens had significantly higher CD-sens values at baseline compared to those who managed with a normal dose, and that the median ratios for anti-Ara h 2 IgE-ab/IgE were significantly higher in the ED group (17%) compared to the ND group (11%). This hypothesis generating study, which had a one-armed study design, provided evidence that the ratio of anti-Ara h 2 IgE-ab/total IgE, as well as basophil CD-sens to peanut, may predict the need for a higher dose of omalizumab in this setting. [DOI] [PubMed] [Google Scholar]