Figure 2. Co-culture of ST2/Rx2Dox murine mesenchymal cells with NBL tumor cells leads to differential gene expression in mesenchymal cells driven towards osteoblastogenesis.
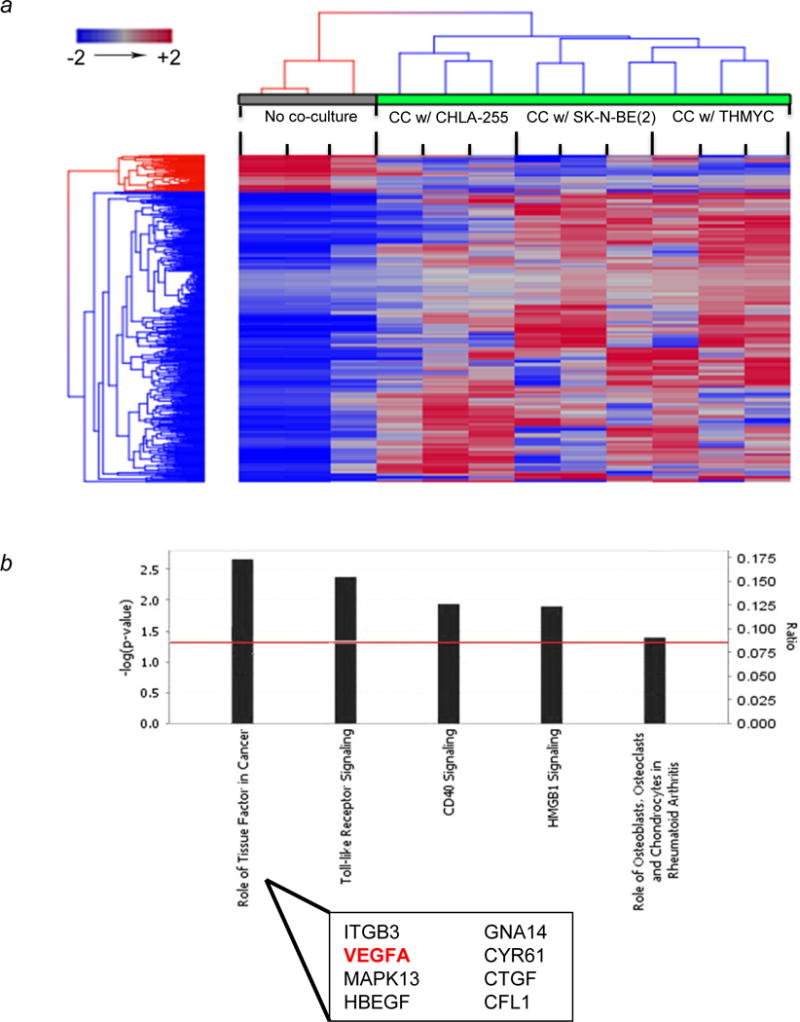
(a) Hierarchical clustering of differentially expressed genes (p<0.05) from ST2/Rx2Dox cells show a distinct pattern between co-culture (green bar) and control (gray bar). ST2/Rx2Dox cells were co-cultured with each of the three NBL cell lines as indicated in Materials and Methods. Euclidean distance and average linkage were used for clustering. Each single column represents technical triplicates. Each group of three columns represents biological triplicates for the same condition. (b) Top canonical pathways involved by differentially expressed genes in co-culture (p<0.05). The significance of canonical pathways was determined by Benjamin-Hocheberg corrected p value (indicated by red line as threshold). The ratio represents the number of differentially expressed genes involved in each canonical pathway. ITGB3, Integrin beta-3 glycoprotein IIIa; MAPK13, Mitogen-activated protein kinase 13; HBEGF, Heparin binding EGF-life growth factor; GNA14, Guanine nucleotide binding protein-14; CYR61, Cysteine-rich angiogenic inducer 61; CTGF, Connective Tissue Growth Factor; CFL1, Cofilin 1.
