Figure 4.
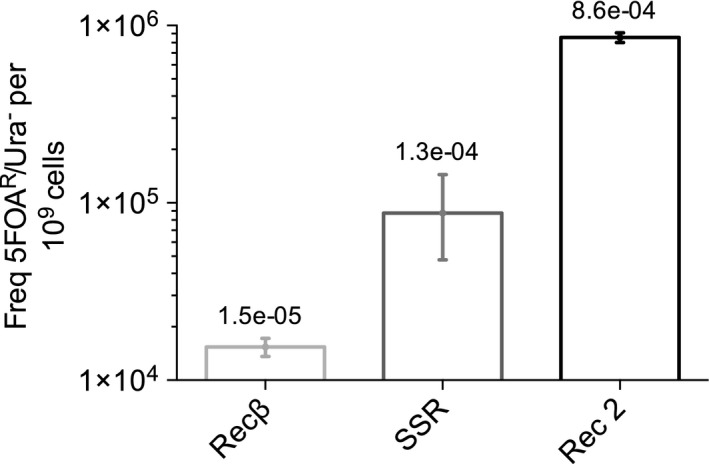
Rec2 maintains recombineering efficiency in entering the LM mutation into the pyrF gene of Pseudomonas putida. P. putida EM42 strains containing pSEVA258‐ssr, pSEVA258‐recβ and pSEVA258‐rec2 were induced with 3MB before electroporation with the LM oligonucleotide (Table S1). After a 6 h recovery period, culture dilutions were plated on selective M9‐Cit‐Ura and M9‐Cit‐Ura‐5FOA solid media. Fifty 5FOAR clones associated with each recombinase were replicated on M9‐Cit and M9‐Cit‐Ura‐5FOA solid media to distinguish authentic pyrF mutants (Ura auxotrophs, no growth on M9‐Cit) from spontaneous 5FOAR colonies (Ura heterotrophs, growth on M9‐Cit). Colony counts were determined and recombineering efficiency calculated (number of authentic pyrF mutants ‐5FOAR/Ura− CFU/109 viable cells). Column graphs of the resulting data were generated with graphpad prism as described in Fig. 2. Each data set represents at least two independent experimental replicates. Recombineering values are shown on a logarithmic scale. Absolute frequency values (mutants per viable cell) are shown above each column.
