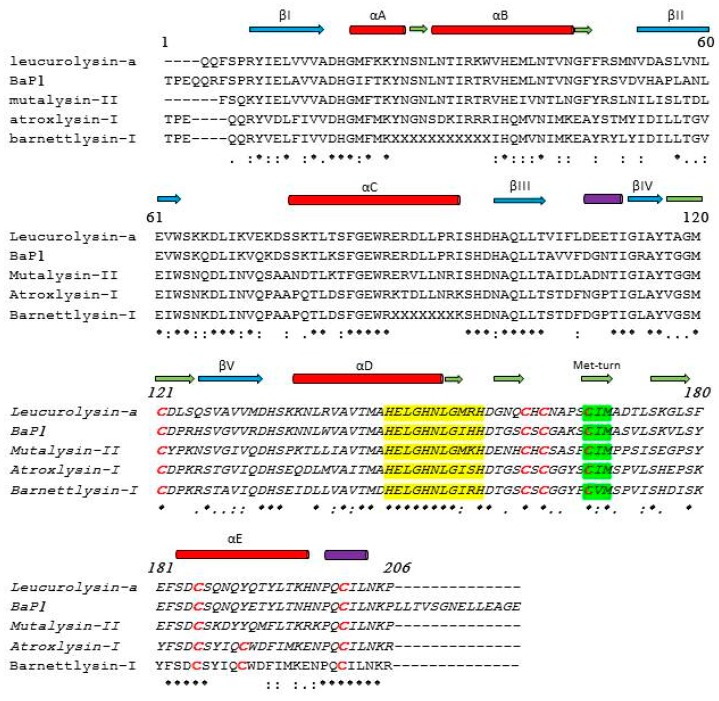
Figure 2.
Sequence comparisons of four P-I class SVMPs. UniProt accession numbers sequences were assigned by using the program ClustalW. Non-hemorrhagic: leuc-a (P84907), mut-II (P22796), bar-I (P86976), and hemorrhagic: atr-I (P85420) and BaP1 (P83512). The sequences of these proteins were determined by the Edman degradation method and the sequences of leuc-a and BaP1 were confirmed by crystallography. Secondary-structure elements were defined by MAFFT V7 (multiple alignment) and PSIPRED V3.3 (predict secondary structure). The blue and dark green arrows indicate the locations of β-strands and turns, respectively, in the crystal structure of leuc-a. The red and purple cylinders represent α-helices and 310 helices, respectively. Cys residues are highlighted in red; (*) identical residues; (:) strongly similar residues; (.) weakly similar residues. The conserved zinc biding motif and the met-turn are highlighted in yellow and bright green, respectively. (-) indicate gaps.