Figure 3.
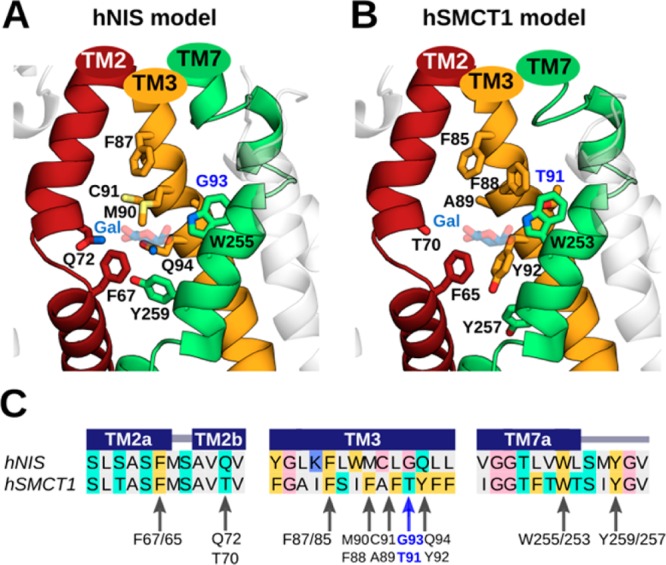
Predicted iodide-binding pockets in hNIS and hSMCT1 and the conservation of associated residues. For the (A) hNIS and (B) hSMCT1 models, these sites involve the transmembrane TM2, TM3, and TM7 segments, which are shown in a cartoon representation and colored as in Figure 1. The nine putative residues coordinating iodide in both models are displayed as sticks colored by atom type (oxygen in red, nitrogen in blue, and sulfur in light yellow). Residues G93/T91 are highlighted in blue. G93 in hNIS was found to be mutated in patients with goitrous hypothyroidism. The galactose (Gal) molecule reported in vSGLT is located near the predicted iodide position in the models and is shown here for reference as transparent blue and red sticks. The predicted iodide binding cavities are displayed with the extracellular side toward the top of the page. (C) Sequence alignment between hNIS and hSMCT1 showing the conservation of residues proposed to coordinate iodide. The alignment is colored according to the chemical properties of the residues as in Figure 1. The secondary structure assignments obtained with PSIPRED (helix) are indicated by dark blue rectangles. Arrows indicate the residues involved in iodide binding according to our prediction.
