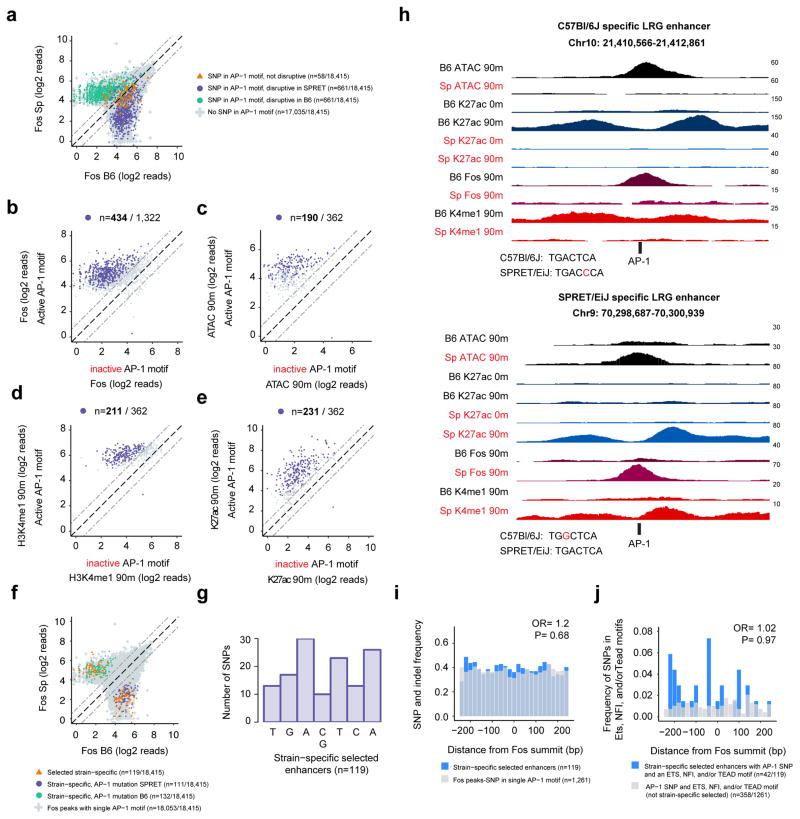
Figure 3. AP-1 TFs are often required for enhancer selection.
a) Fos ChIP-seq signal at all selected enhancers in C57Bl/6J and SPRET/EiJ MEFs that contain a single consensus AP-1 motif and are bound by Fos. SNPs overlapping AP-1 motifs are indicated by their predicted effect on AP-1 binding, b) Fos ChIP-seq signal from each strain for the enhancers identified in (a) at which AP-1 binding would be predicted to be affected by a SNP. Enhancers are classified by whether they contain an active or inactive AP-1 motif, rather than by which strain they come from. Highlighted points indicate enhancers at which Fos binding was significantly strain-specific [FDR<10−6). c–e) Enhancer associated chromatin features for the subset of the enhancers with significant strain-specific Fos binding in (b) that no longer have a Fos peak detected in the strain in which the AP-1 motif is mutated (n=362/434). Highlighted points indicate enhancers at which the chromatin feature was significantly strain-specific (FDR<10−6). f) Fos ChIP-seq signal from each strain. Colored points collectively indicate the 362 enhancers plotted in (c–e). 119 strain-specific enhancers are highlighted (orange triangles) at which the AP-1 SNP leads to both a significant loss of ATAC-seq and H3K4me1 ChIP-seq signal and there is no longer an ATAC-seq peak in the strain in which the AP-1 motif is inactive. g) Histogram showing the location of SNPs in the seven core nucleotides of the AP-1 motif from the 119 strain-specific selected enhancers. h) Representative genome browser tracks for two strain-specific LRG enhancers. AP-1 motifs from each strain are displayed below the enhancer with the SNP highlighted in red. i–j) Total SNP and indel frequency (i) and SNP frequency within Tead/Ets/NFI motifs (j) comparing the 119 strain-specific enhancers (blue) to all other enhancers with a single AP-1 motif that overlaps a SNP (i) (gray) or the subset of these enhancers that contain Tead/Ets/NFI motifs (j).