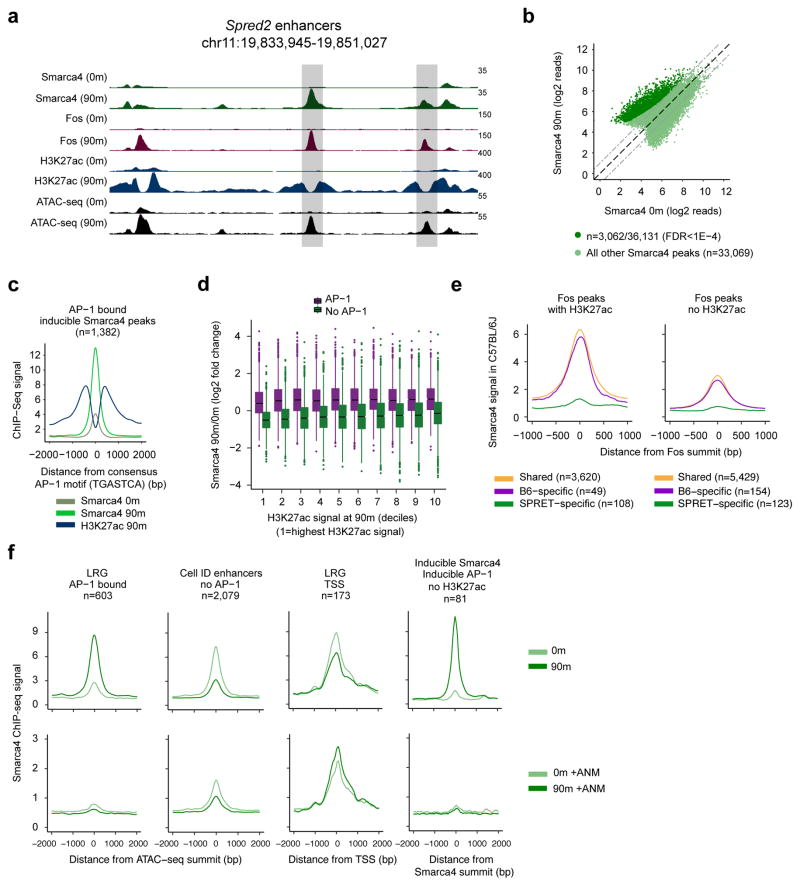
Figure 7. AP-1 TFs are required for BAF recruitment to enhancers.
a) Representative LRG enhancers at the locus of the LRG Spred2. Scale bars indicate normalized read densities for each ChIP-seq and shaded boxes denote LRG enhancers. b) Smarca4 ChIP-seq signal at all Smarca4 peaks before and after stimulation with serum for 90m. Dashed gray lines indicate a 2-fold change. Smarca4 peaks with a significant increase in Smarca4 signal at 90m (n=3,062) compared to 0m are indicated in dark green (FDR<1×10−4). Smarca4 peaks that do not increase significantly at 90m (n=33,069) are indicated in light green. c) Smarca4 and H3K27ac ChIP-seq signal at all inducible Smarca4 peaks bound by AP-1. Smarca4 peaks have been recentered on the closest consensus AP-1 motif within +/−125bp of the Smarca4 peak center. d) Inducible binding of Smarca4 (90m/0m) at ATAC-seq peaks across the genome. ATAC-seq peaks are split into AP-1 bound and not AP-1 bound and binned into deciles according to their levels of H3K27ac ChIP-seq signal [decile 1 =highest H3K27ac signal at 90m; decile 10=lowest H3K27ac signal at 90m). e) Smarca4 ChIP-seq signal from C57Bl/6J MEFs at a set of enhancers that exhibit SPRET/EiJ-specific binding of AP-1 compared to enhancers at which AP-1 binds in both strains. In the left panel, SPRET/EiJ-specific enhancers function as H3K27ac-marked active enhancers in SPRET/EiJ but have an AP-1 point mutation in C57Bl/6J that disrupts binding of Fos (n=108). A similar comparison is shown in the right panel, but instead of focusing on active enhancers in SPRET/EiJ that have lost Fos binding in C57Bl/6J, it displays the Smarca4 ChIP-seq signal at all Fos peaks that are SPRET/EiJ–specific but not at active enhancers (n=123). f) Smarca4 ChIP-seq signal from MEFs in untreated cells (top row) or cells pretreated with the protein synthesis inhibitor anisomycin prior to serum stimulation (bottom row) at different classes of cis-regulatory elements (first three panels) and at Smarca4 peaks at which Smarca4 binding and AP-1 binding are both inducible but that are not at active enhancers marked by H3K27ac.