Figure 6. Chromodomains prevent Chd1 from repositioning a hexasome.
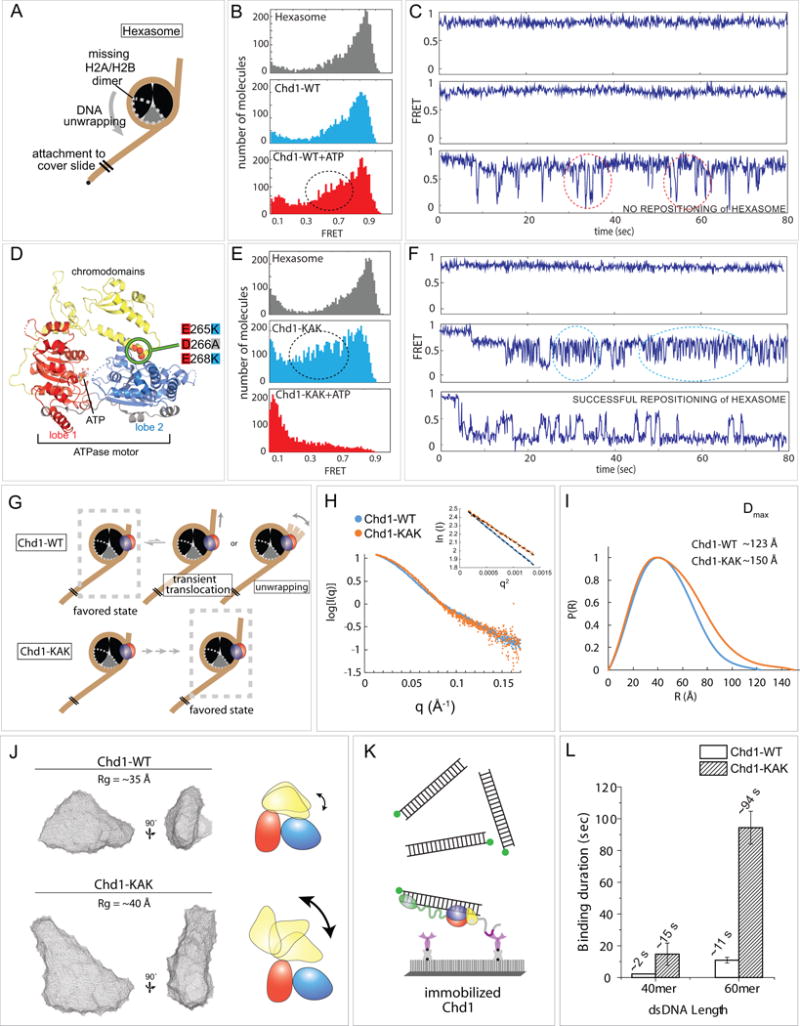
(A) Schematic diagram of hexasome conformation with the dotted gray outline indicating the location of the missing H2A-H2B dimer.
(B) FRET histograms of hexasome alone (gray) and hexasomes plus Chd1 before (light blue) and after ATP addition (red).
(C) Single molecule FRET traces corresponding with conditions in (B).
(D) Crystal structure of the chromodomain-ATPase portion of Chd1 (Hauk et al., 2010), highlighting the location of KAK mutation at the chromo-ATPase interface.
(E) FRET histograms of hexasome alone and hexasomes plus Chd1-KAK mutant, before and after ATP addition.
(F) Single molecule traces corresponding with conditions in (E).
(G) Interpretations of wildtype Chd1 and Chd1-KAK mutant activities on hexasomes.
(H) Small angle X-ray scattering (SAXS) profiles for Chd1-WT and Chd1-KAK proteins consisting of just the chromodomain and ATPase motor. Guinier plot analysis (inset) shows that samples were free from aggregation.
(I) P(R) distributions for SAXS data shown in (H).
(J) Ab initio bead models generated by DAMMIN. Cartoons on the right illustrate possible structural changes associated with the KAK mutation.
(K) Schematic of experiment in which Cy3 labeled dsDNA (40 or 60 bp) was added to surface immobilized Chd1-WT or Chd1-KAK proteins. Note that these constructs contain the DBD.
(L) Binding duration for dsDNA to both proteins (n=500 binding events).
