Fig. 4.
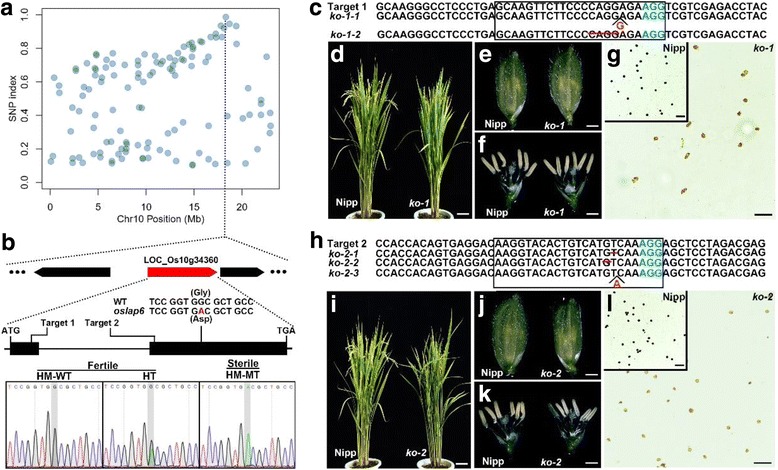
Molecular detection of OsLAP6 and phenotypes of loss-of-function mutants. a The distribution of the SNP sites on chromosome 10. b OsLAP6 gene structure, the mutation site of oslap6 mutant, two independent sgRNA targeting sites of CRISPR/Cas9 system, and the identification of the OsLAP6 gene in the oslap6/9311 F2 population by sequencing. The black boxes indicate the exons. The red characters indicate the SNP mutation in OsLAP6 gene. c and h Sequence alignment of the homologous mutants within the target1 and target2 in the ko-1 and ko-2 T0 plants, respectively. The black frames indicate the target sequences; and the PAM sequences and mutations are in blue and red, respectively. d-g Phenotype of WT and ko-1 in the Nipponbare (Nipp) background. i-l Phenotype of WT and ko-2 in the Nipp background. d and i Plants after heading. e and j Spikelets. f and k Spikelets with the palea and lemma removed. g and l Pollen grains with I2 -KI staining. Scale bars = 5 cm (d and i); 1.5 mm (e and j); 1 mm (f and k); 150 μm (g and l)
