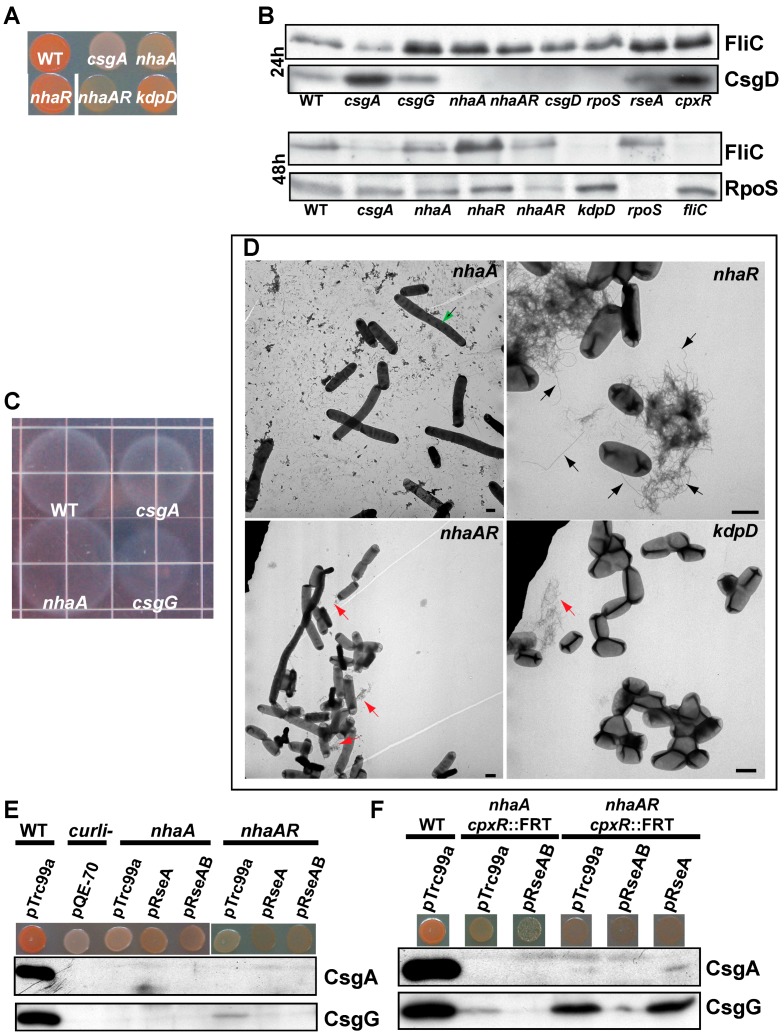
Figure 5.
Sodium antiporter gene nhaA is required for curli production. (A) Strains were grown on CR indicator plates at 26 °C for two days. (B) Whole cell Western blots probed with antibodies to CsgA, CsgG, and FliC at 24 and 48 h. All samples were treated with HFIP; (C) Motility of WT, csgA, csgG, and nhaA strains in 0.2% YESCA motility plates at 26 °C. (D) TEM images from cells grown for 26 °C for two days on YESCA plates. Black arrows indicate flagella. Red arrows indicate curli. Green arrows indicate filamentous cells. Scale bar equals 1 µm. (E) Expression of rseA in trans using pRseA or pRseAB does not rescue curli expression in nhaA or nhaAR strains as detected by CR binding or Western blot probed with antibodies to CsgA or CsgG. (F) Expression of rseA in trans from pRseA or pRseAB does not rescue curli production in nhaA cpxR or nhaAR cpxR double deletions as detected by CR binding or Western blot probed with antibodies to CsgA or CsgG.