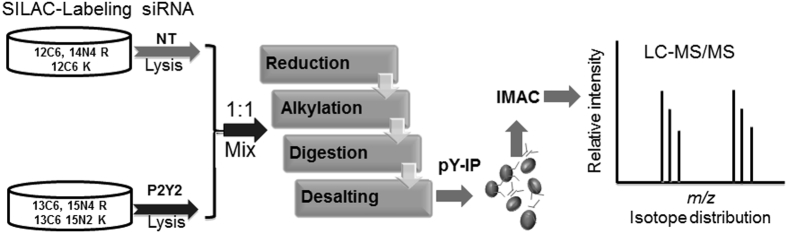
Figure 6.
Schematic for SILAC-based quantitative analysis. The experimental scheme for SILAC-based quantitative phosphoproteome analysis includes culturing HCLEs with SILAC-labeled with heavy- or light-labeled lysine and arginine. The heavy [13C6,15N2 (K), 13C6,15N4 (R)] and light [12C6,14N2(K), 12C6,14N4(R)] isotope-labeled cells were transfected with siRNA directed to P2Y2 receptor and NT (scrambled sequence), respectively. Cells were stimulated with 100 μmol/L UTP for 5 minutes and then lysed, and equal amounts of total protein from each condition were mixed. Lysate was reduced, alkylated and trypsin digested, and subjected to immunoprecipitation using anti-phosphotyrosine antibody. Eluted peptides were further phosphoenriched using IMAC and analyzed using a nanoAcuity UPLC (Waters Corp) coupled through a TriVersa NanoMate (Advion) to an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific).